bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
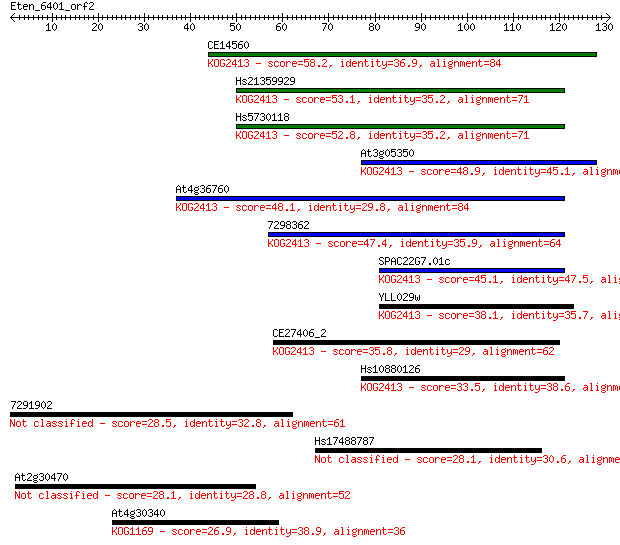
Query= Eten_6401_orf2
Length=130
Score E
Sequences producing significant alignments: (Bits) Value
CE14560 58.2 4e-09
Hs21359929 53.1 1e-07
Hs5730118 52.8 2e-07
At3g05350 48.9 2e-06
At4g36760 48.1 4e-06
7298362 47.4 7e-06
SPAC22G7.01c 45.1 4e-05
YLL029w 38.1 0.004
CE27406_2 35.8 0.020
Hs10880126 33.5 0.10
7291902 28.5 3.5
Hs17488787 28.1 4.3
At2g30470 28.1 4.4
At4g30340 26.9 9.7
> CE14560
Length=616
Score = 58.2 bits (139), Expect = 4e-09, Method: Composition-based stats.
Identities = 31/84 (36%), Positives = 45/84 (53%), Gaps = 1/84 (1%)
Query 44 LQITGALDERRAAWLDPKINLAIHRILADSKRLIVKDTPVSVAKAAKDEKELKGMAEAHE 103
L+ A E +L P+ N AI I+ + + V + V AKA K++ E++GM +H
Sbjct 273 LKAKEASKEPHMVYLTPETNYAIGSIIGEENSM-VDTSLVQTAKATKNDHEMQGMRNSHL 331
Query 104 DDGVALATFFCWFEKQLLANARAT 127
D AL F CW EK+LL+ R T
Sbjct 332 RDSAALVEFLCWLEKELLSGKRYT 355
> Hs21359929
Length=623
Score = 53.1 bits (126), Expect = 1e-07, Method: Composition-based stats.
Identities = 25/71 (35%), Positives = 36/71 (50%), Gaps = 0/71 (0%)
Query 50 LDERRAAWLDPKINLAIHRILADSKRLIVKDTPVSVAKAAKDEKELKGMAEAHEDDGVAL 109
L R W+ K + A+ + R + TP+ +AKA K+ E +GM AH D VAL
Sbjct 282 LSPREKVWVSDKASYAVSETIPKDHRCCMPYTPICIAKAVKNSAESEGMRRAHIKDAVAL 341
Query 110 ATFFCWFEKQL 120
F W EK++
Sbjct 342 CELFNWLEKEV 352
> Hs5730118
Length=623
Score = 52.8 bits (125), Expect = 2e-07, Method: Composition-based stats.
Identities = 25/71 (35%), Positives = 36/71 (50%), Gaps = 0/71 (0%)
Query 50 LDERRAAWLDPKINLAIHRILADSKRLIVKDTPVSVAKAAKDEKELKGMAEAHEDDGVAL 109
L R W+ K + A+ + R + TP+ +AKA K+ E +GM AH D VAL
Sbjct 282 LSPREKVWVSDKASYAVSETIPKDHRCCMPYTPICIAKAVKNSAESEGMRPAHIKDAVAL 341
Query 110 ATFFCWFEKQL 120
F W EK++
Sbjct 342 CELFNWLEKEV 352
> At3g05350
Length=569
Score = 48.9 bits (115), Expect = 2e-06, Method: Composition-based stats.
Identities = 23/51 (45%), Positives = 32/51 (62%), Gaps = 0/51 (0%)
Query 77 IVKDTPVSVAKAAKDEKELKGMAEAHEDDGVALATFFCWFEKQLLANARAT 127
I +P+S AKA K++ ELKGM +H D ALA F+ W E+++ NA T
Sbjct 237 IYMQSPISWAKAIKNDAELKGMKNSHLRDAAALAHFWAWLEEEVHKNANLT 287
> At4g36760
Length=634
Score = 48.1 bits (113), Expect = 4e-06, Method: Composition-based stats.
Identities = 25/85 (29%), Positives = 50/85 (58%), Gaps = 4/85 (4%)
Query 37 RHEPRSKLQITGALDERRAAWLDP-KINLAIHRILADSKRLIVKDTPVSVAKAAKDEKEL 95
+HE ++I D+ W+DP A++ L D+++++++ +P+S++KA K+ EL
Sbjct 264 QHEAAKDMEIDS--DQPDRLWVDPASCCYALYSKL-DAEKVLLQPSPISLSKALKNPVEL 320
Query 96 KGMAEAHEDDGVALATFFCWFEKQL 120
+G+ AH DG A+ + W + Q+
Sbjct 321 EGIKNAHVRDGAAVVQYLVWLDNQM 345
> 7298362
Length=613
Score = 47.4 bits (111), Expect = 7e-06, Method: Composition-based stats.
Identities = 23/64 (35%), Positives = 37/64 (57%), Gaps = 1/64 (1%)
Query 57 WLDPKINLAIHRILADSKRLIVKDTPVSVAKAAKDEKELKGMAEAHEDDGVALATFFCWF 116
W+ P + + ++ S+R I + TP+ V KA K++ E+ G +H DGVAL +F W
Sbjct 282 WIAPTSSYYLTALIPKSRR-IQEVTPICVLKAIKNDVEIAGFINSHIRDGVALCQYFAWL 340
Query 117 EKQL 120
E Q+
Sbjct 341 EDQV 344
> SPAC22G7.01c
Length=389
Score = 45.1 bits (105), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 19/40 (47%), Positives = 26/40 (65%), Gaps = 0/40 (0%)
Query 81 TPVSVAKAAKDEKELKGMAEAHEDDGVALATFFCWFEKQL 120
+P+S AK K++ ELKGM E H DG AL +F W ++ L
Sbjct 294 SPISQAKGIKNDAELKGMKECHIRDGCALVEYFAWLDEYL 333
> YLL029w
Length=749
Score = 38.1 bits (87), Expect = 0.004, Method: Composition-based stats.
Identities = 15/42 (35%), Positives = 25/42 (59%), Gaps = 0/42 (0%)
Query 81 TPVSVAKAAKDEKELKGMAEAHEDDGVALATFFCWFEKQLLA 122
+P+ V K+ K++ E+K +A D V L +F W E+QL+
Sbjct 447 SPIDVLKSIKNDIEIKNAHKAQVKDAVCLVQYFAWLEQQLVG 488
> CE27406_2
Length=608
Score = 35.8 bits (81), Expect = 0.020, Method: Composition-based stats.
Identities = 18/62 (29%), Positives = 33/62 (53%), Gaps = 1/62 (1%)
Query 58 LDPKINLAIHRILADSKRLIVKDTPVSVAKAAKDEKELKGMAEAHEDDGVALATFFCWFE 117
+ P+ N I R++ + +I + + K K+ +LKGM ++ D +A+ F C FE
Sbjct 281 ISPETNYLIGRLIGEDHSMI-DPSIMERIKKIKNTDQLKGMRASNLRDSIAIVEFLCKFE 339
Query 118 KQ 119
K+
Sbjct 340 KE 341
> Hs10880126
Length=674
Score = 33.5 bits (75), Expect = 0.10, Method: Composition-based stats.
Identities = 17/46 (36%), Positives = 25/46 (54%), Gaps = 2/46 (4%)
Query 77 IVKDT--PVSVAKAAKDEKELKGMAEAHEDDGVALATFFCWFEKQL 120
+V DT PV + KA K+ KE + +H D VA+ + W EK +
Sbjct 342 LVTDTYSPVMMTKAVKNSKEQALLKASHVRDAVAVIRYLVWLEKNV 387
> 7291902
Length=498
Score = 28.5 bits (62), Expect = 3.5, Method: Composition-based stats.
Identities = 20/63 (31%), Positives = 30/63 (47%), Gaps = 2/63 (3%)
Query 1 QLLQREHQQLPEDLQQQTHGGLLGAVDWLLAGGDETR-HEPRSKLQITGALD-ERRAAWL 58
QL Q Q+LP L+++ + A +WL A R + SK + G LD E A+
Sbjct 170 QLSQYRAQRLPRSLKKEHENNFISASEWLEARSKRGRSSDSVSKYEQLGELDYESDWAYA 229
Query 59 DPK 61
P+
Sbjct 230 GPE 232
> Hs17488787
Length=362
Score = 28.1 bits (61), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 15/49 (30%), Positives = 25/49 (51%), Gaps = 10/49 (20%)
Query 67 HRILADSKRLIVKDTPVSVAKAAKDEKELKGMAEAHEDDGVALATFFCW 115
HR+LAD+ RL T + + + D++ +D + + TFFCW
Sbjct 299 HRLLADAHRLYAVQTNLWLYQWMDDDR----------NDIMFVTTFFCW 337
> At2g30470
Length=780
Score = 28.1 bits (61), Expect = 4.4, Method: Composition-based stats.
Identities = 15/52 (28%), Positives = 28/52 (53%), Gaps = 8/52 (15%)
Query 2 LLQREHQQLPEDLQQQTHGGLLGAVDWLLAGGDETRHEPRSKLQITGALDER 53
L+ +E +PE+L +T+GG + G D TR + + + + GA ++R
Sbjct 425 LIPKELNGMPENLNSETNGGRI--------GDDPTRVKEKKRTRTIGAKNKR 468
> At4g30340
Length=490
Score = 26.9 bits (58), Expect = 9.7, Method: Composition-based stats.
Identities = 14/36 (38%), Positives = 19/36 (52%), Gaps = 0/36 (0%)
Query 23 LGAVDWLLAGGDETRHEPRSKLQITGALDERRAAWL 58
LG +D L A GDE E R K++I A + W+
Sbjct 140 LGCLDTLAAKGDECARECREKIRIMVAGGDGTVGWV 175
Lambda K H
0.317 0.133 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1246445644
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40