bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
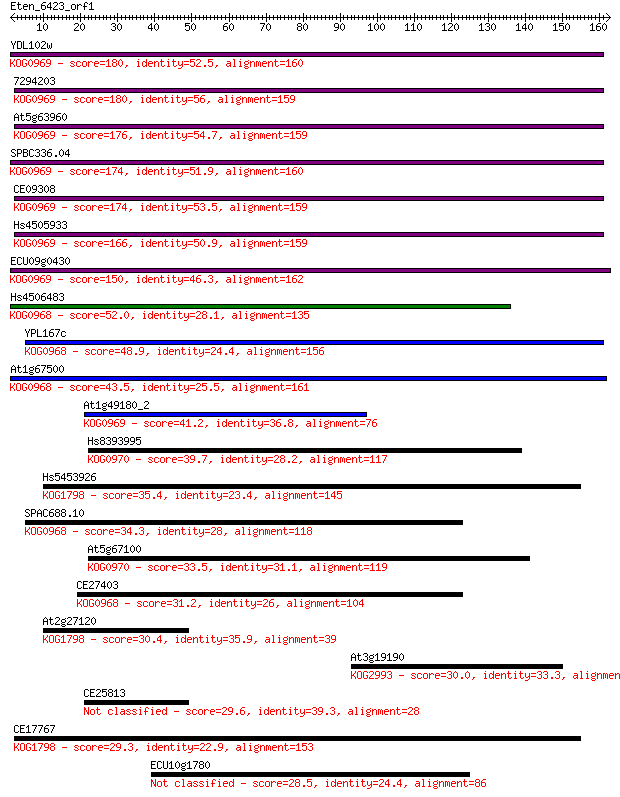
Query= Eten_6423_orf1
Length=162
Score E
Sequences producing significant alignments: (Bits) Value
YDL102w 180 1e-45
7294203 180 1e-45
At5g63960 176 2e-44
SPBC336.04 174 5e-44
CE09308 174 9e-44
Hs4505933 166 3e-41
ECU09g0430 150 1e-36
Hs4506483 52.0 5e-07
YPL167c 48.9 4e-06
At1g67500 43.5 2e-04
At1g49180_2 41.2 0.001
Hs8393995 39.7 0.003
Hs5453926 35.4 0.049
SPAC688.10 34.3 0.098
At5g67100 33.5 0.18
CE27403 31.2 0.86
At2g27120 30.4 1.7
At3g19190 30.0 2.0
CE25813 29.6 2.6
CE17767 29.3 3.1
ECU10g1780 28.5 6.7
> YDL102w
Length=1097
Score = 180 bits (457), Expect = 1e-45, Method: Compositional matrix adjust.
Identities = 84/160 (52%), Positives = 112/160 (70%), Gaps = 0/160 (0%)
Query 1 AGALVVWFDDESEMLKKWAEFVREVDPDFLTGYNCINFDLCYLLTRASALKAEGVASLSR 60
G+++ E EML W F+ +VDPD + GYN NFD+ YLL RA ALK R
Sbjct 369 TGSMIFSHATEEEMLSNWRNFIIKVDPDVIIGYNTTNFDIPYLLNRAKALKVNDFPYFGR 428
Query 61 LKSIESRISDTRFSSRALGTHDNKEIPMEGRVQFDLLELIRRDHKLKSYSLNYVSSEFLK 120
LK+++ I ++ FSS+A GT + K + ++GR+Q DLL+ I+R++KL+SY+LN VS+ FL
Sbjct 429 LKTVKQEIKESVFSSKAYGTRETKNVNIDGRLQLDLLQFIQREYKLRSYTLNAVSAHFLG 488
Query 121 EQKEDVHYSMIGDLFRGSPTTRRRIAVYCLKDALLPLRLL 160
EQKEDVHYS+I DL G TRRR+AVYCLKDA LPLRL+
Sbjct 489 EQKEDVHYSIISDLQNGDSETRRRLAVYCLKDAYLPLRLM 528
> 7294203
Length=1092
Score = 180 bits (456), Expect = 1e-45, Method: Compositional matrix adjust.
Identities = 89/159 (55%), Positives = 113/159 (71%), Gaps = 0/159 (0%)
Query 2 GALVVWFDDESEMLKKWAEFVREVDPDFLTGYNCINFDLCYLLTRASALKAEGVASLSRL 61
G+ V+ D E++ML KW+ FVREVDPD LTGYN NFD YLL RA+ LK L R+
Sbjct 349 GSQVLCHDKETQMLDKWSAFVREVDPDILTGYNINNFDFPYLLNRAAHLKVRNFEYLGRI 408
Query 62 KSIESRISDTRFSSRALGTHDNKEIPMEGRVQFDLLELIRRDHKLKSYSLNYVSSEFLKE 121
K+I S I + S+ +G +N+ + EGRV FDLL ++ RD+KL+SY+LN VS FL+E
Sbjct 409 KNIRSVIKEQMLQSKQMGRRENQYVNFEGRVPFDLLFVLLRDYKLRSYTLNAVSYHFLQE 468
Query 122 QKEDVHYSMIGDLFRGSPTTRRRIAVYCLKDALLPLRLL 160
QKEDVH+S+I DL G TRRR+A+YCLKDA LPLRLL
Sbjct 469 QKEDVHHSIITDLQNGDEQTRRRLAMYCLKDAYLPLRLL 507
> At5g63960
Length=1081
Score = 176 bits (446), Expect = 2e-44, Method: Compositional matrix adjust.
Identities = 87/159 (54%), Positives = 110/159 (69%), Gaps = 0/159 (0%)
Query 2 GALVVWFDDESEMLKKWAEFVREVDPDFLTGYNCINFDLCYLLTRASALKAEGVASLSRL 61
G V+ F+ E E+L W + +R+VDPD + GYN FDL YL+ RA+ L E L R+
Sbjct 359 GVDVMSFETEREVLLAWRDLIRDVDPDIIIGYNICKFDLPYLIERAATLGIEEFPLLGRV 418
Query 62 KSIESRISDTRFSSRALGTHDNKEIPMEGRVQFDLLELIRRDHKLKSYSLNYVSSEFLKE 121
K+ R+ D+ FSSR G ++KE +EGR QFDL++ I RDHKL SYSLN VS+ FL E
Sbjct 419 KNSRVRVRDSTFSSRQQGIRESKETTIEGRFQFDLIQAIHRDHKLSSYSLNSVSAHFLSE 478
Query 122 QKEDVHYSMIGDLFRGSPTTRRRIAVYCLKDALLPLRLL 160
QKEDVH+S+I DL G+ TRRR+AVYCLKDA LP RLL
Sbjct 479 QKEDVHHSIITDLQNGNAETRRRLAVYCLKDAYLPQRLL 517
> SPBC336.04
Length=1086
Score = 174 bits (442), Expect = 5e-44, Method: Compositional matrix adjust.
Identities = 83/160 (51%), Positives = 113/160 (70%), Gaps = 0/160 (0%)
Query 1 AGALVVWFDDESEMLKKWAEFVREVDPDFLTGYNCINFDLCYLLTRASALKAEGVASLSR 60
G V F +++EML W++FVR+VDPD L GYN NFD+ YLL RA +L+ L R
Sbjct 348 VGTQVYEFQNQAEMLSSWSKFVRDVDPDVLIGYNICNFDIPYLLDRAKSLRIHNFPLLGR 407
Query 61 LKSIESRISDTRFSSRALGTHDNKEIPMEGRVQFDLLELIRRDHKLKSYSLNYVSSEFLK 120
+ + S +T FSS+A GT ++K + GR+Q D+L++++RD KL+SYSLN V S+FL
Sbjct 408 IHNFFSVAKETTFSSKAYGTRESKTTSIPGRLQLDMLQVMQRDFKLRSYSLNAVCSQFLG 467
Query 121 EQKEDVHYSMIGDLFRGSPTTRRRIAVYCLKDALLPLRLL 160
EQKEDVHYS+I DL G+ +RRR+A+YCLKDA LP RL+
Sbjct 468 EQKEDVHYSIITDLQNGTADSRRRLAIYCLKDAYLPQRLM 507
> CE09308
Length=1081
Score = 174 bits (440), Expect = 9e-44, Method: Compositional matrix adjust.
Identities = 85/159 (53%), Positives = 113/159 (71%), Gaps = 0/159 (0%)
Query 2 GALVVWFDDESEMLKKWAEFVREVDPDFLTGYNCINFDLCYLLTRASALKAEGVASLSRL 61
G+ ++ +E +L+KWAEFVREVDPD +TGYN +NFDL Y+L RA L V+ L R
Sbjct 335 GSNIIQCVNEKVLLEKWAEFVREVDPDIITGYNILNFDLPYILDRAKVLSLPQVSHLGRQ 394
Query 62 KSIESRISDTRFSSRALGTHDNKEIPMEGRVQFDLLELIRRDHKLKSYSLNYVSSEFLKE 121
K S + D SS+ +G+ NK I + GR+ FD+L+++ RD+KL+SY+LN VS +FL E
Sbjct 395 KEKGSVVRDAAISSKQMGSRVNKSIDIHGRIIFDVLQVVLRDYKLRSYTLNSVSYQFLSE 454
Query 122 QKEDVHYSMIGDLFRGSPTTRRRIAVYCLKDALLPLRLL 160
QKEDV +++I DL RG TRRR+A YCLKDA LPLRLL
Sbjct 455 QKEDVEHNIIPDLQRGDEQTRRRLAQYCLKDAYLPLRLL 493
> Hs4505933
Length=1107
Score = 166 bits (419), Expect = 3e-41, Method: Compositional matrix adjust.
Identities = 81/159 (50%), Positives = 111/159 (69%), Gaps = 0/159 (0%)
Query 2 GALVVWFDDESEMLKKWAEFVREVDPDFLTGYNCINFDLCYLLTRASALKAEGVASLSRL 61
GA V ++ E ++L+ W+ F+R +DPD +TGYN NFDL YL++RA LK + L R+
Sbjct 365 GAKVQSYEKEEDLLQAWSTFIRIMDPDVITGYNIQNFDLPYLISRAQTLKVQTFPFLGRV 424
Query 62 KSIESRISDTRFSSRALGTHDNKEIPMEGRVQFDLLELIRRDHKLKSYSLNYVSSEFLKE 121
+ S I D+ F S+ G D K + M GRVQ D+L+++ R++KL+S++LN VS FL E
Sbjct 425 AGLCSNIRDSSFQSKQTGRRDTKVVSMVGRVQMDMLQVLLREYKLRSHTLNAVSFHFLGE 484
Query 122 QKEDVHYSMIGDLFRGSPTTRRRIAVYCLKDALLPLRLL 160
QKEDV +S+I DL G+ TRRR+AVYCLKDA LPLRLL
Sbjct 485 QKEDVQHSIITDLQNGNDQTRRRLAVYCLKDAYLPLRLL 523
> ECU09g0430
Length=974
Score = 150 bits (378), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 75/162 (46%), Positives = 103/162 (63%), Gaps = 1/162 (0%)
Query 1 AGALVVWFDDESEMLKKWAEFVREVDPDFLTGYNCINFDLCYLLTRASALKAEGVASLSR 60
G V W++ E E+L+ W ++ E+DPD + GYN FD+ Y+L+R L E + L R
Sbjct 247 PGVNVHWYETEKELLESWKKYFMELDPDVIVGYNVKGFDIPYILSRGEILGIESFSVLGR 306
Query 61 LKSIESRISDTRFSSRALGTHDNKEIPMEGRVQFDLLELIRRDHKLKSYSLNYVSSEFLK 120
+ ++R DT SS G+ E+ ++GR+ D++ +IRRD KL+SYSLN VS FLK
Sbjct 307 SRK-KARTRDTTMSSNMFGSITTTEVEIDGRLIIDMMVVIRRDFKLRSYSLNSVSIHFLK 365
Query 121 EQKEDVHYSMIGDLFRGSPTTRRRIAVYCLKDALLPLRLLTN 162
EQKEDV YS IG+L + TRRRIA YCL+D +LPLRL
Sbjct 366 EQKEDVPYSSIGELQSKNKDTRRRIASYCLRDTVLPLRLFNT 407
> Hs4506483
Length=3052
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 38/137 (27%), Positives = 64/137 (46%), Gaps = 5/137 (3%)
Query 1 AGALVVWFDDESEMLKKWAEFVREVDPDFLTGYNCINFDLCYLLTRASALKAEGVASLSR 60
G V + DE + + A ++ DPD L GY YLL RA+AL + +SR
Sbjct 2294 TGLEVTYAADEKALFHEIANIIKRYDPDILLGYEIQMHSWGYLLQRAAALSIDLCRMISR 2353
Query 61 LK--SIESRISDTRFSSRALGTHDNKEIPMEGRVQFDLLELIRRDHKLKSYSLNYVSSEF 118
+ IE+R + R G++ EI + GR+ +L ++R + L +Y+ VS
Sbjct 2354 VPDDKIENRFAAER---DEYGSYTMSEINIVGRITLNLWRIMRNEVALTNYTFENVSFHV 2410
Query 119 LKEQKEDVHYSMIGDLF 135
L ++ + ++ D F
Sbjct 2411 LHQRFPLFTFRVLSDWF 2427
> YPL167c
Length=1504
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 38/160 (23%), Positives = 80/160 (50%), Gaps = 10/160 (6%)
Query 5 VVWFDDESEMLKKWAEFVREVDPDFLTGYNCINFDLCYLLTRASAL-KAEGVASLSRLK- 62
V++++ E EM + + V +DPD L+G+ NF Y++ R + + + V L+R+K
Sbjct 739 VMFYESEFEMFEALTDLVLLLDPDILSGFEIHNFSWGYIIERCQKIHQFDIVRELARVKC 798
Query 63 SIESRISDTRFSSRALGTHDNKEIPMEGRVQFDLLELIRRDHKLKSYSLNYVSSEFLKEQ 122
I++++SDT + + G I + GR ++ +R D L Y++ + L ++
Sbjct 799 QIKTKLSDTWGYAHSSG------IMITGRHMINIWRALRSDVNLTQYTIESAAFNILHKR 852
Query 123 KEDVHYSMIGDLF--RGSPTTRRRIAVYCLKDALLPLRLL 160
+ + +++ + S T + + Y L A + ++LL
Sbjct 853 LPHFSFESLTNMWNAKKSTTELKTVLNYWLSRAQINIQLL 892
> At1g67500
Length=1894
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 41/187 (21%), Positives = 83/187 (44%), Gaps = 28/187 (14%)
Query 1 AGALVVWFDDESEMLKKWAEFVREVDPDFLTGYNCINFDLCYLLTRASALKAEGVASLSR 60
+G + F +E ++ + + E + + DPD L G++ + +L RA+ L + ++SR
Sbjct 1097 SGCKLSVFLEERQLFRYFIETLCKWDPDVLLGWDIQGGSIGFLAERAAQLGIRFLNNISR 1156
Query 61 LKSIESRISDTRFSSRALGTH-----------DNKEIPME--------------GRVQFD 95
S + ++ + R LG + +E+ +E GR+ +
Sbjct 1157 TPSPTT--TNNSDNKRKLGNNLLPDPLVANPAQVEEVVIEDEWGRTHASGVHVGGRIVLN 1214
Query 96 LLELIRRDHKLKSYSLNYVSSEFLKEQKEDVHYSMIGDLFRGSPT-TRRRIAVYCLKDAL 154
LIR + KL Y++ VS L+++ + Y ++ + F P R R Y ++ A
Sbjct 1215 AWRLIRGEVKLNMYTIEAVSEAVLRQKVPSIPYKVLTEWFSSGPAGARYRCIEYVIRRAN 1274
Query 155 LPLRLLT 161
L L +++
Sbjct 1275 LNLEIMS 1281
> At1g49180_2
Length=167
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 40/79 (50%), Gaps = 11/79 (13%)
Query 21 FVREVDPDFLTGYNCINFDLCYLL---TRASALKAEGVASLSRLKSIESRISDTRFSSRA 77
++++ PD CI D+C L A+ L E L R+K+ + DT FSSR
Sbjct 3 ILQQMHPD------CI--DVCSRLLSINPAATLGIEDFPFLGRIKNSRVWVKDTTFSSRQ 54
Query 78 LGTHDNKEIPMEGRVQFDL 96
GT + K + GR+QFDL
Sbjct 55 HGTRERKVATINGRLQFDL 73
> Hs8393995
Length=1462
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 33/118 (27%), Positives = 55/118 (46%), Gaps = 9/118 (7%)
Query 22 VREVDPDFLTGYNCINFDLCYLLTRASALKAEGVASLSRLKSIESRISDTRFSSRA-LGT 80
V ++DPD + G+N F+L LL R + KA + + RLK R + + R+ G
Sbjct 626 VHKIDPDIIVGHNIYGFELEVLLQRINVCKAPHWSKIGRLK----RSNMPKLGGRSGFGE 681
Query 81 HDNKEIPMEGRVQFDLLELIRRDHKLKSYSLNYVSSEFLKEQKEDVHYSMIGDLFRGS 138
+ M V+ ELIR KSY L+ + + LK ++ + I +++ S
Sbjct 682 RNATCGRMICDVEISAKELIR----CKSYHLSELVQQILKTERVVIPMENIQNMYSES 735
> Hs5453926
Length=2286
Score = 35.4 bits (80), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 34/145 (23%), Positives = 60/145 (41%), Gaps = 19/145 (13%)
Query 10 DESEMLKKWAEFVREVDPDFLTGYNCINFDLCYLLTRASALKAEGVASLSRLKSIESRIS 69
DE+ ++++W E V+E P + YN FD ++ RA+ V LS + I
Sbjct 339 DEAHLIQRWFEHVQETKPTIMVTYNGDFFDWPFVEARAA------VHGLSMQQEI----- 387
Query 70 DTRFSSRALGTHDNKEIPMEGRVQFDLLELIRRDHKLKSYSLNYVSSEFLKEQKEDVHYS 129
F + G + + + D L ++RD L S N ++ K + V
Sbjct 388 --GFQKDSQGEYKAPQC-----IHMDCLRWVKRDSYLPVGSHNLKAAAKAKLGYDPVELD 440
Query 130 MIGDLFRGSPTTRRRIAVYCLKDAL 154
D+ R + + +A Y + DA+
Sbjct 441 P-EDMCRMATEQPQTLATYSVSDAV 464
> SPAC688.10
Length=1480
Score = 34.3 bits (77), Expect = 0.098, Method: Compositional matrix adjust.
Identities = 33/121 (27%), Positives = 53/121 (43%), Gaps = 7/121 (5%)
Query 5 VVWFDDESEMLKKWAEFVREVDPDFLTGYNCINFDLCYLLTRASA-LKAEGVASLSRLKS 63
V+ + E E++ + R++DP + GY N YL+ RAS + LSRLK
Sbjct 717 VLVVNSELELINEVIGLNRQLDPTIVCGYEVHNSSWGYLIERASYRFNYDLPEQLSRLKC 776
Query 64 IESRISDTRFSSR--ALGTHDNKEIPMEGRVQFDLLELIRRDHKLKSYSLNYVSSEFLKE 121
S F+ + A I + GR ++ ++R + L +YSL V K+
Sbjct 777 ----TSKANFAKKENAWKYTTTSSINIVGRHVLNIWRILRGEVNLLNYSLENVVLNIFKK 832
Query 122 Q 122
Q
Sbjct 833 Q 833
> At5g67100
Length=1492
Score = 33.5 bits (75), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 37/123 (30%), Positives = 58/123 (47%), Gaps = 7/123 (5%)
Query 22 VREVDPDFLTGYNCINFDLCYLLTRASALKAEGV--ASLSRLK-SIESRISDTRFSSRAL 78
+ ++D D L G+N FDL LL RA A K + + + RLK S ++ S+
Sbjct 660 LNKLDSDILVGHNISGFDLDVLLQRAQACKVQSSMWSKIGRLKRSFMPKLKGN--SNYGS 717
Query 79 GTHDNKEIPMEGRVQFDLLELIRRD-HKLKSYSLNYVSSEFLKEQKEDVHYSMIGDLFRG 137
G + GR+ D +L RD K SYSL +S L ++++ + I +F+
Sbjct 718 GATPGLMSCIAGRLLCD-TDLCSRDLLKEVSYSLTDLSKTQLNRDRKEIAPNDIPKMFQS 776
Query 138 SPT 140
S T
Sbjct 777 SKT 779
> CE27403
Length=1154
Score = 31.2 bits (69), Expect = 0.86, Method: Compositional matrix adjust.
Identities = 27/110 (24%), Positives = 49/110 (44%), Gaps = 20/110 (18%)
Query 19 AEFVREVDPDFLTGYNCINFDLCYLLTRASALKAEGVASLSRLKSIESRISDTRFSSRAL 78
A+++ + D D + GY I + R+K + SRIS R A
Sbjct 442 AKWIVQYDVDVMIGYETIRLSWGFF--------------FRRIKLLGSRISMDRALIDAY 487
Query 79 GTH---DNKEI---PMEGRVQFDLLELIRRDHKLKSYSLNYVSSEFLKEQ 122
H D++EI P +GR+ + +++R D L++Y L + L+++
Sbjct 488 EDHIEVDDQEITVAPPKGRLLVSVWKVVRSDLALRNYDLGSAVANVLRKK 537
> At2g27120
Length=2154
Score = 30.4 bits (67), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 10 DESEMLKKWAEFVREVDPDFLTGYNCINFDLCYLLTRAS 48
+E E+L+KW ++E+ P YN FD ++ RAS
Sbjct 303 NEVELLRKWFSHMQELKPGIYVTYNGDFFDWPFIERRAS 341
> At3g19190
Length=1814
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 19/58 (32%), Positives = 25/58 (43%), Gaps = 1/58 (1%)
Query 93 QFDLLEL-IRRDHKLKSYSLNYVSSEFLKEQKEDVHYSMIGDLFRGSPTTRRRIAVYC 149
Q +EL ++ H Y V L E EDV + I L +G PT R A+Y
Sbjct 1556 QMQGIELQLKHVHAAGIYGWGNVCETILGEWLEDVSQNQIHQLLKGIPTVRSLSALYA 1613
> CE25813
Length=924
Score = 29.6 bits (65), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 11/28 (39%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 21 FVREVDPDFLTGYNCINFDLCYLLTRAS 48
F++ +D D+L +N +N L +L TR+S
Sbjct 119 FIKHIDYDYLEEFNNLNIQLLHLATRSS 146
> CE17767
Length=2144
Score = 29.3 bits (64), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 35/156 (22%), Positives = 68/156 (43%), Gaps = 24/156 (15%)
Query 2 GALVVWFD-DESEMLKKWAEFVREVDPDFLTGYNCINFDLCYLLTRASALKAEGVASLSR 60
G VW + DE+ +++K+ + +V P+ + YN FD ++ RA K G
Sbjct 314 GEFTVWNEKDEAALIRKFFDHFLQVRPNIVVTYNGDFFDWPFVEARA---KIRGF----- 365
Query 61 LKSIESRISDTRFSSRALGTHDNKEIPMEGRVQFDLLELIRRDHKLK--SYSLNYVSSEF 118
++E I FS + + ++ + D ++RD L S +L V+
Sbjct 366 --NMEREIG---FSKDSADEYKSRNC-----IHMDAFRWVKRDSYLPVGSQNLKAVTKAK 415
Query 119 LKEQKEDVHYSMIGDLFRGSPTTRRRIAVYCLKDAL 154
L+ +V ++ + R P +++A Y + DA+
Sbjct 416 LRYDPVEVEPELMCKMAREQP---QQLANYSVSDAV 448
> ECU10g1780
Length=423
Score = 28.5 bits (62), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 21/87 (24%), Positives = 43/87 (49%), Gaps = 18/87 (20%)
Query 39 DLCYLLTRASALKAEGVASLSR-LKSIESRISDTRFSSRALGTHDNKEIPMEGRVQFDLL 97
++C++ + L E L+R ++++ESR+ + R+ R + +L
Sbjct 331 EVCHVKGSGAVLLVE--KQLTRCIEALESRVFNKRYDVRHTDS---------------VL 373
Query 98 ELIRRDHKLKSYSLNYVSSEFLKEQKE 124
ELI+++ K Y +Y+ FLK++ E
Sbjct 374 ELIKQNLKFSKYDFSYLMEHFLKKRSE 400
Lambda K H
0.322 0.137 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2244926132
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40