bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6436_orf2
Length=152
Score E
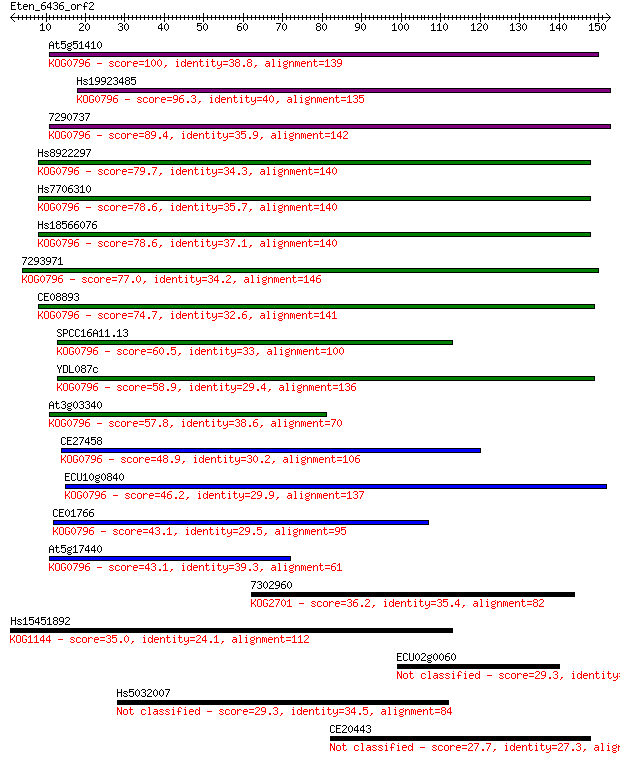
Sequences producing significant alignments: (Bits) Value
At5g51410 100 1e-21
Hs19923485 96.3 2e-20
7290737 89.4 2e-18
Hs8922297 79.7 2e-15
Hs7706310 78.6 4e-15
Hs18566076 78.6 4e-15
7293971 77.0 1e-14
CE08893 74.7 6e-14
SPCC16A11.13 60.5 1e-09
YDL087c 58.9 3e-09
At3g03340 57.8 8e-09
CE27458 48.9 3e-06
ECU10g0840 46.2 3e-05
CE01766 43.1 2e-04
At5g17440 43.1 2e-04
7302960 36.2 0.027
Hs15451892 35.0 0.053
ECU02g0060 29.3 2.8
Hs5032007 29.3 2.9
CE20443 27.7 9.4
> At5g51410
Length=334
Score = 100 bits (248), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 54/144 (37%), Positives = 82/144 (56%), Gaps = 5/144 (3%)
Query 11 MNEMRATLDSLMGRDRNEYGANAKKGASFKAD--SVCKFYLLDFCPHDLFPNTRVDLGPC 68
M+ RA LD LMG RN + K D VC FY++ FCPHDLF NT+ DLG C
Sbjct 1 MDAQRALLDELMGAARNLTDEERRGFKEVKWDDREVCAFYMVRFCPHDLFVNTKSDLGAC 60
Query 69 DREHRADLKVAFETDPEREFYQALFETEFARYLQRLVDQMENRIKRVQQRIDANNQPV-- 126
R H LK +FE P + Y FE E A++ ++LV+ ++ +++R ++R+ +PV
Sbjct 61 SRIHDPKLKESFENSPRHDSYVPKFEAELAQFCEKLVNDLDRKVRRGRERLAQEVEPVPP 120
Query 127 -ELSKENEERVNSMNAEISELLKQ 149
LS E E+++ + ++ LL+Q
Sbjct 121 PSLSAEKAEQLSVLEEKVKNLLEQ 144
> Hs19923485
Length=432
Score = 96.3 bits (238), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 54/140 (38%), Positives = 81/140 (57%), Gaps = 7/140 (5%)
Query 18 LDSLMGRDRNEYGANAKKGASFKADSVCKFYLLDFCPHDLFPNTRVDLGPCDREHRADLK 77
LD LMGRDRN + + +SVCK+YL FCP +LF NTR DLGPC++ H +L+
Sbjct 8 LDELMGRDRNLAPDEKRSNVRWDHESVCKYYLCGFCPAELFTNTRSDLGPCEKIHDENLR 67
Query 78 VAFETDPEREFYQALFETEFARYLQRLVDQMENRIKRVQQRID-ANNQ----PVELSKEN 132
+E F + +E +F RYLQ L+ ++E RI+R R+ + NQ + +N
Sbjct 68 KQYEKSS--RFMKVGYERDFLRYLQSLLAEVERRIRRGHARLALSQNQQSSGAAGPTGKN 125
Query 133 EERVNSMNAEISELLKQQDE 152
EE++ + +I LL+Q +E
Sbjct 126 EEKIQVLTDKIDVLLQQIEE 145
> 7290737
Length=414
Score = 89.4 bits (220), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 51/147 (34%), Positives = 83/147 (56%), Gaps = 7/147 (4%)
Query 11 MNEMRATLDSLMGRDRNEYGANAKKGASFKADSVCKFYLLDFCPHDLFPNTRVDLGPCDR 70
++ R LD LMGR+RN + + A +++ C+FY + FCPHDLF NTR DLGPC R
Sbjct 2 VDAARQMLDELMGRNRNLHPSEAGAKVNWEDPEFCQFYNVKFCPHDLFINTRADLGPCAR 61
Query 71 EHRADLKVAFETDPEREFYQALFETEFARYLQRLVDQMENRIKRVQQRI-----DANNQP 125
H + + +E D + +E EF R+ ++ ++ +I++ +QR+ D N P
Sbjct 62 IHDEEARHLYE-DARPSQRKRQYEDEFLRFCNVMLHDVDRKIQKGKQRLLLMQRDQPNVP 120
Query 126 VELSKENEERVNSMNAEISELLKQQDE 152
LS+ E+ N + A I++LL + +E
Sbjct 121 APLSRHQEQLAN-LTARINKLLSEAEE 146
> Hs8922297
Length=325
Score = 79.7 bits (195), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 48/143 (33%), Positives = 78/143 (54%), Gaps = 9/143 (6%)
Query 8 FSKMNEMRATLDSLMGRDRNEYGANAKKGASFKADSVCKFYLLDFCPHDLFPNTRVDLGP 67
S +MRA LD LMG R+ G ++ F D VCK +LLD CPHD+ TR+DLG
Sbjct 1 MSAQAQMRALLDQLMGTARD--GDETRQRVKFTDDRVCKSHLLDCCPHDILAGTRMDLGE 58
Query 68 CDREHRADLKVAFE-TDPEREFYQALFETEFARYLQRLVDQMENRIKRVQQRIDANNQPV 126
C + H L+ +E ER+ + FE + +L+ + + + R + ++R+ + +
Sbjct 59 CTKIHDLALRADYEIASKERDLF---FELDAMDHLESFIAECDRRTELAKKRLAETQEEI 115
Query 127 --ELSKENEERVNSMNAEISELL 147
E+S + E+V+ +N EI +LL
Sbjct 116 SAEVSAKA-EKVHELNEEIGKLL 137
> Hs7706310
Length=402
Score = 78.6 bits (192), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 50/144 (34%), Positives = 77/144 (53%), Gaps = 11/144 (7%)
Query 8 FSKMNEMRATLDSLMGRDRNEYGANAKKGASFKADSVCKFYLLDFCPHDLFPNTRVDLGP 67
S +MRA LD LMG R+ G ++ F D VCK +LL+ CPHD+ TR+DLG
Sbjct 1 MSAQAQMRAMLDQLMGTSRD--GDTTRQRIKFSDDRVCKSHLLNCCPHDVLSGTRMDLGE 58
Query 68 CDREHRADLKVAFE-TDPEREFYQALFETEFARYLQRLVDQMENRIKRVQQRIDANNQPV 126
C + H L+ +E E++F+ FE + +LQ + + R + ++R+ +
Sbjct 59 CLKVHDLALRADYEIASKEQDFF---FELDAMDHLQSFIADCDRRTEVAKKRLAETQE-- 113
Query 127 ELSKE---NEERVNSMNAEISELL 147
E+S E ERV+ +N EI +LL
Sbjct 114 EISAEVAAKAERVHELNEEIGKLL 137
> Hs18566076
Length=392
Score = 78.6 bits (192), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 52/144 (36%), Positives = 78/144 (54%), Gaps = 11/144 (7%)
Query 8 FSKMNEMRATLDSLMGRDRNEYGANAKKGASFKADSVCKFYLLDFCPHDLFPNTRVDLGP 67
S +MRA LD LMG R+ G ++ F D VCK +LL+ CPHD+ TR+DLG
Sbjct 1 MSAQAQMRAMLDQLMGTSRD--GDTTRQRIKFSDDRVCKSHLLNCCPHDVLSGTRMDLGE 58
Query 68 CDREHRADLKVAFE-TDPEREFYQALFETEFARYLQRLVDQMENRIKRVQQRIDANNQPV 126
C + H L+ +E E++F+ FE + +LQ + + R + ++R+ A Q
Sbjct 59 CLKVHDLALRADYEIASKEQDFF---FELDAMDHLQSFIADCDRRTEVAKKRL-AETQE- 113
Query 127 ELSKE---NEERVNSMNAEISELL 147
E+S E ERV+ +N EI +LL
Sbjct 114 EISAEVAAKAERVHELNEEIGKLL 137
> 7293971
Length=420
Score = 77.0 bits (188), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 50/150 (33%), Positives = 78/150 (52%), Gaps = 16/150 (10%)
Query 4 PYLSFSKMNEMRATLDSLMGRDRNEYGANAKKGASFKADSVCKFYLLDFCPHDLFPNTRV 63
P S ++MRA LD LMG RN ++ F VCK +LLD CPHD+ +TR+
Sbjct 4 PSNKMSATDQMRAMLDQLMGTTRN----GDERQLKFSDPRVCKSFLLDCCPHDILASTRM 59
Query 64 DLGPCDREH----RADLKVAFETDPEREFYQALFETEFARYLQRLVDQMENRIKRVQQRI 119
DLG C + H RAD + A +T R++Y ++ E +LQ + + R +QR+
Sbjct 60 DLGECPKVHDLAFRADYESAAKT---RDYY---YDIEAMEHLQAFIADCDRRTDSAKQRL 113
Query 120 DANNQPVELSKENEERVNSMNAEISELLKQ 149
+ EL+ E E+ N+++ E+ K+
Sbjct 114 KETQE--ELTAEVAEKANAVHGLAEEIGKK 141
> CE08893
Length=369
Score = 74.7 bits (182), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 46/147 (31%), Positives = 77/147 (52%), Gaps = 10/147 (6%)
Query 8 FSKMNEMRATLDSLMGRDRN-EYGANAKKGASFKADSVCKFYLLDFCPHDLFPNTRVDLG 66
S N M A LD LMG RN E G + K +F +C ++L+ FCPHD+F NT+ DLG
Sbjct 1 MSARNAMAAMLDELMGPKRNVELGKDTK--VTFDDPDICPYFLVGFCPHDMFINTKADLG 58
Query 67 PCDREHRADLKVAFETDPEREFYQALFETEFARYLQRLVDQMENRIKRVQQRIDANNQPV 126
C H +L+ + P E+ Q FE R+L +L + + RI++ + ++ +
Sbjct 59 ACQLVHDDNLRRLYPESP--EYGQLGFERRLMRFLVQLDEDNQRRIRKNKDKLSGMDDSG 116
Query 127 ELSKENEER-----VNSMNAEISELLK 148
+ E E+R +NS++ + +++
Sbjct 117 KRKLEEEKRQVQLEINSIDDVLKNMIR 143
> SPCC16A11.13
Length=264
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 33/100 (33%), Positives = 51/100 (51%), Gaps = 4/100 (4%)
Query 13 EMRATLDSLMGRDRNEYGANAKKGASFKADSVCKFYLLDFCPHDLFPNTRVDLGPCDREH 72
E R ++ LMG + + + ++ F VC+ +L CPHD+F NT++DLGPC + H
Sbjct 4 EQRKIIEQLMGSNLSNF--TSRGLVHFTDRKVCRSFLCGICPHDIFTNTKMDLGPCPKIH 61
Query 73 RADLKVAFETDPEREFYQALFETEFARYLQRLVDQMENRI 112
LK +E Y +E ++ L+R VD RI
Sbjct 62 SDKLKSDYERASYSHDYG--YEWDYLEDLERHVDDCNKRI 99
> YDL087c
Length=261
Score = 58.9 bits (141), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 40/146 (27%), Positives = 69/146 (47%), Gaps = 10/146 (6%)
Query 13 EMRATLDSLMGRD---RNEYGANAKKGASFKADSVCKFYLLDFCPHDLFPNTRVDLGPCD 69
E R ++ LMGRD R+ ++ K+ +CK YL+ CP+DLF T+ LG C
Sbjct 10 EQRKLVEQLMGRDFSFRHNRYSHQKRDLGLHDPKICKSYLVGECPYDLFQGTKQSLGKCP 69
Query 70 REHRADLKVAFETDPEREFYQALFETEFARYLQRLVDQMENRIKRVQQRIDANNQP-VEL 128
+ H K+ +E + ++ FE E+ L R V++ +I Q + + +++
Sbjct 70 QMHLTKHKIQYEREVKQGKTFPEFEREYLAILSRFVNECNGQISVALQNLKHTAEERMKI 129
Query 129 SKENEE------RVNSMNAEISELLK 148
+ EE R+ M EI L++
Sbjct 130 QQVTEELDVLDVRIGLMGQEIDSLIR 155
> At3g03340
Length=385
Score = 57.8 bits (138), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 27/70 (38%), Positives = 39/70 (55%), Gaps = 2/70 (2%)
Query 11 MNEMRATLDSLMGRDRNEYGANAKKGASFKADSVCKFYLLDFCPHDLFPNTRVDLGPCDR 70
M+ +R LD LMG +RN G + + VC+ YL CPHDLF T++D+GPC +
Sbjct 1 MDAIRKQLDVLMGANRN--GDVQEVNRKYYDRDVCRLYLSGLCPHDLFQLTKMDMGPCPK 58
Query 71 EHRADLKVAF 80
H L+ +
Sbjct 59 VHSLQLRKEY 68
> CE27458
Length=339
Score = 48.9 bits (115), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 32/109 (29%), Positives = 59/109 (54%), Gaps = 8/109 (7%)
Query 14 MRATLDSLMGRDRNEYGANAKKGASFKAD--SVCKFYLLDFCPHDLFPNTRVDLGPCD-R 70
M L+ LMG R+ AN + + D +VC +L+ FC HD+F NT+ DLG C
Sbjct 5 MAQMLNELMGSQRD---ANPGERREIRYDDPNVCTDFLVGFCTHDIFRNTKNDLGFCKYT 61
Query 71 EHRADLKVAFETDPEREFYQALFETEFARYLQRLVDQMENRIKRVQQRI 119
H +LK +++ ++ ++ FE F ++R+ + + +I++ + R+
Sbjct 62 THDENLKNSYKNSDKK--WRMGFEKRFLERIRRIHEDVRRKIQKHEDRL 108
> ECU10g0840
Length=245
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 41/139 (29%), Positives = 64/139 (46%), Gaps = 8/139 (5%)
Query 15 RATLDSLMGRDRNEYGANAKKGASFKADSVCKFYLLDFCPHDLFPNTRVDLGPCDREHRA 74
R L+ L+G +R+ Y + + S K D VC + L+ FCP +LF NTR +G C
Sbjct 6 REVLNMLLGPERDTY--DPCRPTSTKKD-VCIYMLVSFCPFELFRNTRRSIGKCRYTSHE 62
Query 75 DLKVAFETDPEREFYQALFETEFARYLQRLVDQMENRIKRVQQRIDANNQPVELS--KEN 132
+ A RE + +E EF R L +V +++ I+ + ++ P KE
Sbjct 63 EYYKAEYNRNGRERAEE-YEWEFVRLLVEIVLSVQDGIR--ARGLEDRTDPALFGKIKEK 119
Query 133 EERVNSMNAEISELLKQQD 151
EE N + + EL D
Sbjct 120 EELFNRVYESVGELGMSGD 138
> CE01766
Length=313
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 28/98 (28%), Positives = 50/98 (51%), Gaps = 6/98 (6%)
Query 12 NEMRATLDSLMGRDRNEYGANAKK-GASFKADSVCKFYLLDFCPHDLFPNTRV-DLGPCD 69
++MR + LMG +++ N +K F SVC+ +LL CPHD+ P++R+ ++ C
Sbjct 3 DQMRDMIAQLMG---SQHVDNKEKPSMPFDHHSVCRAFLLGVCPHDMVPDSRLQNVVSCR 59
Query 70 REHRADLKVAFE-TDPEREFYQALFETEFARYLQRLVD 106
+ H K +E E++ + + E + LVD
Sbjct 60 KVHEPAHKADYERAQKEKDHFYDVDAFEIIEHAVHLVD 97
> At5g17440
Length=381
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 24/61 (39%), Positives = 31/61 (50%), Gaps = 2/61 (3%)
Query 11 MNEMRATLDSLMGRDRNEYGANAKKGASFKADSVCKFYLLDFCPHDLFPNTRVDLGPCDR 70
M+ MR LD LMG +RN G + + VC+ YL CPH+LF T + DR
Sbjct 1 MDAMRKQLDVLMGANRN--GDVTEVNRKYYDRDVCRLYLSGLCPHELFQLTAKGVDNYDR 58
Query 71 E 71
E
Sbjct 59 E 59
> 7302960
Length=595
Score = 36.2 bits (82), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 47/90 (52%), Gaps = 13/90 (14%)
Query 62 RVDLGPCDRE----HRADLKVAFETDPEREFYQALFETEFARYLQRLVDQMENRIKRV-- 115
R+DL P R+ H A+ K+ ETD Q L + L+ ME++I+R
Sbjct 265 RIDLDPTARQELQQHYAEFKLEMETDA-----QELKSQNQQKRLEAAKIAMEHKIRRYCK 319
Query 116 --QQRIDANNQPVELSKENEERVNSMNAEI 143
+Q DA N+ E +KE E+++N++N E+
Sbjct 320 ENEQLQDAENEQKEKTKEVEDQLNTLNTEL 349
> Hs15451892
Length=1220
Score = 35.0 bits (79), Expect = 0.053, Method: Compositional matrix adjust.
Identities = 27/112 (24%), Positives = 47/112 (41%), Gaps = 9/112 (8%)
Query 1 DGFPYLSFSKMNEMRATLDSLMGRDRNEYGANAKKGASFKADSVCKFYLLDFCPHDLFPN 60
D P L ++E++ TL+++ ++ Y A S +A LL+F P
Sbjct 954 DEIPVLKDELIHELKQTLNAIKLEEKGVY-VQASTLGSLEA-------LLEFLKTSEVPY 1005
Query 61 TRVDLGPCDREHRADLKVAFETDPEREFYQALFETEFARYLQRLVDQMENRI 112
+++GP ++ V E DP+ A F+ R Q + D + RI
Sbjct 1006 AGINIGPVHKKDVMKASVMLEHDPQYAVILA-FDVRIERDAQEMADSLGVRI 1056
> ECU02g0060
Length=602
Score = 29.3 bits (64), Expect = 2.8, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 24/41 (58%), Gaps = 5/41 (12%)
Query 99 RYLQRLVDQMENRIKRVQQRIDANNQPVELSKENEERVNSM 139
R+ QRLV ++E +IK + +N P KE EER+N +
Sbjct 159 RFGQRLVREVEVKIKEM-----SNEMPSSEKKEKEERLNQI 194
> Hs5032007
Length=322
Score = 29.3 bits (64), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 29/89 (32%), Positives = 38/89 (42%), Gaps = 7/89 (7%)
Query 28 EYGANAKKGASFKADSVC---KFYLLDFCPH--DLFPNTRVDLGPCDREHRADLKVAFET 82
E GA K + SV + YL DF H L P+ DL E R LK F
Sbjct 90 EVGAGGNKSRLTLSMSVAVEFRDYLGDFIEHYAQLGPSQPPDLAQAQDEPRRALKSEFLV 149
Query 83 DPEREFYQALFETEFARYLQRLVDQMENR 111
R++Y L E + R+L+ + Q NR
Sbjct 150 RENRKYYMDLKENQRGRFLR--IRQTVNR 176
> CE20443
Length=727
Score = 27.7 bits (60), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 18/73 (24%), Positives = 36/73 (49%), Gaps = 7/73 (9%)
Query 82 TDPEREFYQALF-------ETEFARYLQRLVDQMENRIKRVQQRIDANNQPVELSKENEE 134
T+P ++F+Q F E ++ Y + LV + +I R+ I N + V+ + E
Sbjct 98 TEPSKQFFQEQFVKEWQYHEIDYTPYFETLVPRFREKIARLLDEIVKNPEEVDDDVQEEL 157
Query 135 RVNSMNAEISELL 147
R + + + ++ LL
Sbjct 158 RWHLLFSPVTTLL 170
Lambda K H
0.318 0.134 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1925034518
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40