bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6519_orf4
Length=163
Score E
Sequences producing significant alignments: (Bits) Value
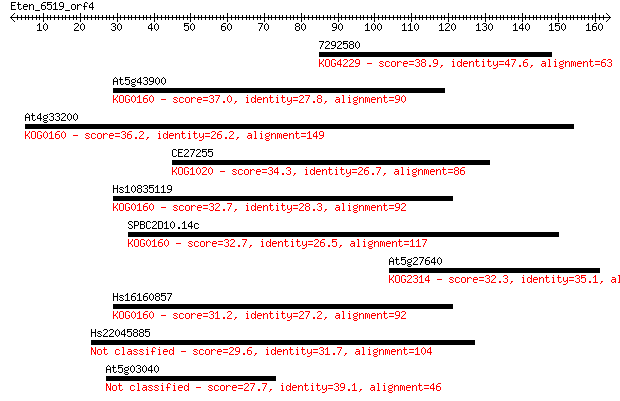
7292580 38.9 0.005
At5g43900 37.0 0.017
At4g33200 36.2 0.026
CE27255 34.3 0.12
Hs10835119 32.7 0.33
SPBC2D10.14c 32.7 0.34
At5g27640 32.3 0.39
Hs16160857 31.2 0.99
Hs22045885 29.6 2.9
At5g03040 27.7 9.3
> 7292580
Length=2424
Score = 38.9 bits (89), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 30/63 (47%), Positives = 40/63 (63%), Gaps = 15/63 (23%)
Query 85 RRQLMQQIHSRRQAAATKIQARWRARAQQLRFQQERLTRCLLRVRQLAATEIQRVWKGRC 144
RRQL +RRQAAAT++QARWR + Q R++ R+R+ A T QR+W+GR
Sbjct 860 RRQL-----ARRQAAATRLQARWRGQRAQQRYE---------RLRKGALT-AQRLWRGRQ 904
Query 145 ARR 147
ARR
Sbjct 905 ARR 907
> At5g43900
Length=1505
Score = 37.0 bits (84), Expect = 0.017, Method: Composition-based stats.
Identities = 25/90 (27%), Positives = 43/90 (47%), Gaps = 14/90 (15%)
Query 29 AASTIQRFIRGYFTRLRYQVALVWRRLLQQQQQQEQQAATRIQCCWRRRKAQQEAKRRQL 88
AA IQ + RGY RL Y R+L ++AA QC WR + A+ E ++ ++
Sbjct 833 AAIIIQTWCRGYLARLHY------RKL--------KKAAITTQCAWRSKVARGELRKLKM 878
Query 89 MQQIHSRRQAAATKIQARWRARAQQLRFQQ 118
+ QAA K++ + +L+ ++
Sbjct 879 AARETGALQAAKNKLEKQVEELTWRLQLEK 908
> At4g33200
Length=1374
Score = 36.2 bits (82), Expect = 0.026, Method: Composition-based stats.
Identities = 39/154 (25%), Positives = 65/154 (42%), Gaps = 36/154 (23%)
Query 5 RRDCGAPLLPPSRIQ--LLQCMIAD--TAASTIQRFIRGYFTRLRYQVALVWRRLLQQQQ 60
RR+ A +L ++ L +C +AA +Q IR TRL++
Sbjct 774 RRNAAAAVLVQKHVRRWLSRCAFVKLVSAAIVLQSCIRADSTRLKFS------------H 821
Query 61 QQEQQAATRIQCCWRRRKAQQEAKRRQLMQQIHSRRQAAATKIQARWRARAQQLRFQQER 120
Q+E +AA+ IQ WR K + + RQ ++ IQ RWR + + F+
Sbjct 822 QKEHRAASLIQAHWRIHKFRSAFRHRQ----------SSIIAIQCRWRQKLAKREFR--- 868
Query 121 LTRCLLRVRQLA-ATEIQRVWKGRCARRALDEEW 153
+++Q+A R+ K + +R D EW
Sbjct 869 ------KLKQVANEAGALRLAKTKLEKRLEDLEW 896
> CE27255
Length=2227
Score = 34.3 bits (77), Expect = 0.12, Method: Composition-based stats.
Identities = 23/86 (26%), Positives = 40/86 (46%), Gaps = 0/86 (0%)
Query 45 RYQVALVWRRLLQQQQQQEQQAATRIQCCWRRRKAQQEAKRRQLMQQIHSRRQAAATKIQ 104
R ++A R L +QQ+ +EQ R+ R +A + R++ I +++QA K+Q
Sbjct 223 RLRIAEEKRLLEEQQRLREQMERERLAEIKRLEEAARLEDERRIAADIEAQKQAMLQKMQ 282
Query 105 ARWRARAQQLRFQQERLTRCLLRVRQ 130
A ++ Q+ L RV Q
Sbjct 283 AEQNKHIAEVERQRSELEERFARVSQ 308
> Hs10835119
Length=1855
Score = 32.7 bits (73), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 26/103 (25%), Positives = 45/103 (43%), Gaps = 26/103 (25%)
Query 29 AASTIQRFIRGYFTRLRY-------QVALVWRR----LLQQQQQQEQQAATRIQCCWRRR 77
A +Q ++RG+ R RY + ++ +R L + ++ A +QCC+RR
Sbjct 840 ATIVLQSYLRGFLARNRYRKILRGHKAVIIQKRVRGWLARTHYKRSMHAIIYLQCCFRRM 899
Query 78 KAQQEAKRRQLMQQIHSRRQAAATKIQARWRARAQQLRFQQER 120
A++E K+ KI+AR R ++LR E
Sbjct 900 MAKRELKK---------------LKIEARSVERYKKLRIGMEN 927
> SPBC2D10.14c
Length=1471
Score = 32.7 bits (73), Expect = 0.34, Method: Composition-based stats.
Identities = 31/128 (24%), Positives = 54/128 (42%), Gaps = 29/128 (22%)
Query 33 IQRFIRGYFTRLRYQ-----------VALVWRRLLQQQQQQEQQAATRIQCCWRRRKAQQ 81
+Q IRG+FTR YQ V + W + ++++ ++AA IQ WR
Sbjct 759 LQSAIRGFFTRKEYQRTVKFIIKLQSVIMGWLTRQRFEREKIERAAILIQAHWR-----S 813
Query 82 EAKRRQLMQQIHSRRQAAATKIQARWRARAQQLRFQQERLTRCLLRVRQLAATEIQRVWK 141
+R++ + I A IQ+ R R+ E +R+ +AT + + W+
Sbjct 814 YIQRKRYLSLIK-----CAIVIQSIVRKNIAYSRYINE--------LRESSATLLAKFWR 860
Query 142 GRCARRAL 149
AR+
Sbjct 861 AYNARKTF 868
> At5g27640
Length=738
Score = 32.3 bits (72), Expect = 0.39, Method: Composition-based stats.
Identities = 20/64 (31%), Positives = 30/64 (46%), Gaps = 7/64 (10%)
Query 104 QARWRARAQQLRF--QQERLTRCLLRVRQLAATEIQRVW-----KGRCARRALDEEWEAW 156
Q WR R ++E + + L + + E Q V + R R+AL EEWE W
Sbjct 627 QLAWRPRPPSFLTAEKEEEIAKTLKKYSKKYEAEDQDVSLLLSEQDREKRKALKEEWEKW 686
Query 157 VVKW 160
V++W
Sbjct 687 VMQW 690
> Hs16160857
Length=1855
Score = 31.2 bits (69), Expect = 0.99, Method: Compositional matrix adjust.
Identities = 25/103 (24%), Positives = 44/103 (42%), Gaps = 26/103 (25%)
Query 29 AASTIQRFIRGYFTRLRY-------QVALVWRR----LLQQQQQQEQQAATRIQCCWRRR 77
A +Q ++RG+ R RY + ++ +R L + ++ A +QCC+RR
Sbjct 840 ATIVLQSYLRGFLARNRYRKILREHKAVIIQKRVRGWLARTHYKRSMHAIIYLQCCFRRM 899
Query 78 KAQQEAKRRQLMQQIHSRRQAAATKIQARWRARAQQLRFQQER 120
A++E K+ KI+AR R ++L E
Sbjct 900 MAKRELKK---------------LKIEARSVERYKKLHIGMEN 927
> Hs22045885
Length=146
Score = 29.6 bits (65), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 33/107 (30%), Positives = 51/107 (47%), Gaps = 20/107 (18%)
Query 23 CMIADTAASTIQRFIRGYFTRLRYQVALVWRRLLQQQQQQEQQAATR---IQCCWRRRKA 79
C+ + A IQ + RG R R LLQ AA R IQC WR +A
Sbjct 22 CLNLEKTAIKIQSWWRGNMVR---------RTLLQ--------AALRAWVIQCWWRSMQA 64
Query 80 QQEAKRRQLMQQIHSRRQAAATKIQARWRARAQQLRFQQERLTRCLL 126
+ +RR+L ++++ ++ A K+QA+ R + RF Q R C++
Sbjct 65 KMLEQRRRLALRLYTCQEWAVVKVQAQVRMWQARRRFLQARQAACII 111
> At5g03040
Length=445
Score = 27.7 bits (60), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 18/48 (37%), Positives = 25/48 (52%), Gaps = 2/48 (4%)
Query 27 DTAASTIQRFIRGYFTR--LRYQVALVWRRLLQQQQQQEQQAATRIQC 72
+ AA IQ RGY R LR LV +LL + ++QAA ++C
Sbjct 114 EAAAILIQTIFRGYLARRALRAMRGLVRLKLLMEGSVVKRQAANTLKC 161
Lambda K H
0.328 0.131 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2281134618
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40