bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6590_orf1
Length=119
Score E
Sequences producing significant alignments: (Bits) Value
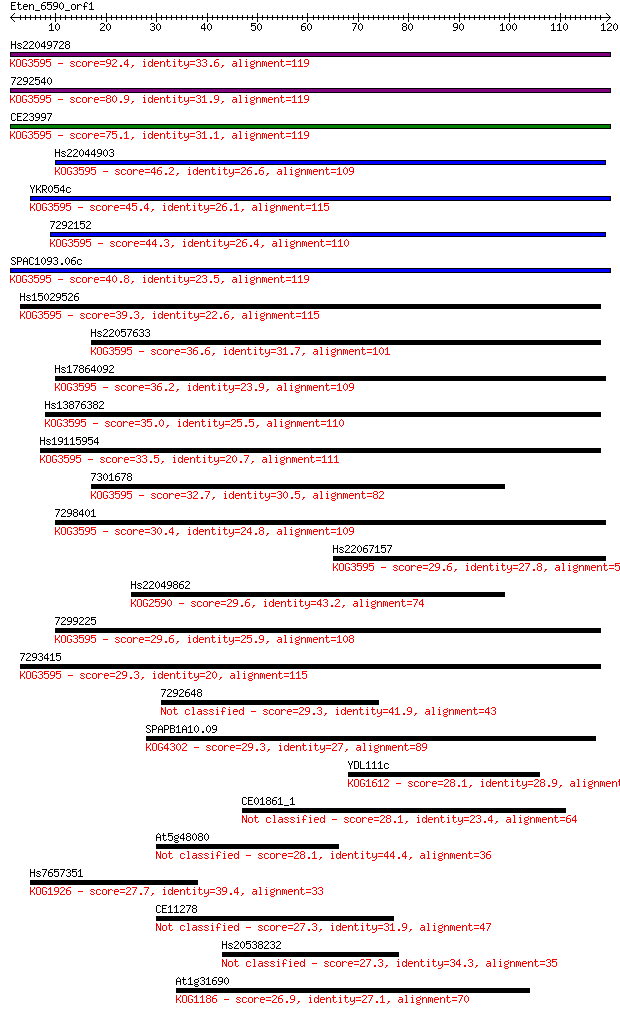
Hs22049728 92.4 2e-19
7292540 80.9 5e-16
CE23997 75.1 3e-14
Hs22044903 46.2 1e-05
YKR054c 45.4 2e-05
7292152 44.3 6e-05
SPAC1093.06c 40.8 6e-04
Hs15029526 39.3 0.002
Hs22057633 36.6 0.010
Hs17864092 36.2 0.014
Hs13876382 35.0 0.038
Hs19115954 33.5 0.11
7301678 32.7 0.16
7298401 30.4 0.84
Hs22067157 29.6 1.3
Hs22049862 29.6 1.4
7299225 29.6 1.5
7293415 29.3 1.6
7292648 29.3 1.9
SPAPB1A10.09 29.3 2.1
YDL111c 28.1 3.9
CE01861_1 28.1 3.9
At5g48080 28.1 4.1
Hs7657351 27.7 4.7
CE11278 27.3 6.5
Hs20538232 27.3 7.4
At1g31690 26.9 8.2
> Hs22049728
Length=2274
Score = 92.4 bits (228), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 40/119 (33%), Positives = 72/119 (60%), Gaps = 7/119 (5%)
Query 1 IAQFITKWMILSILWGMGGSLSLAARSAFSEEVARFCDLPLPSQLASSDADGQTLLDFEP 60
+ ++I ++++ +ILW + G L R+ E + R +PLP+ A ++D+E
Sbjct 114 LERYIQRYLVYAILWSLSGDSRLKMRAELGEYIRRITTVPLPT------APNIPIIDYEV 167
Query 61 SIDDGQWHHWKSRVKQVEIDTQQVKDATLVIQTVDTLRHRDVVEGWLEEKRPFILCGPP 119
SI G+W W+++V Q+E++T +V +V+ T+DT+RH ++ WL E +P +LCGPP
Sbjct 168 SIS-GEWSPWQAKVPQIEVETHKVAAPDVVVPTLDTVRHEALLYTWLAEHKPLVLCGPP 225
> 7292540
Length=4680
Score = 80.9 bits (198), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 38/119 (31%), Positives = 64/119 (53%), Gaps = 7/119 (5%)
Query 1 IAQFITKWMILSILWGMGGSLSLAARSAFSEEVARFCDLPLPSQLASSDADGQTLLDFEP 60
+ +I K ++ S+LW G L R + V +PLP A G ++D+E
Sbjct 2471 LEHYIPKALVYSVLWSFAGDAKLKVRIDLGDFVRSVTTVPLPG------AAGAPIIDYEV 2524
Query 61 SIDDGQWHHWKSRVKQVEIDTQQVKDATLVIQTVDTLRHRDVVEGWLEEKRPFILCGPP 119
++ G W W ++V +E++T +V +V+ T+DT+RH ++ WL E +P +LCGPP
Sbjct 2525 NMS-GDWVPWSNKVPVIEVETHKVASPDIVVPTLDTVRHESLLYTWLAEHKPLVLCGPP 2582
> CE23997
Length=4568
Score = 75.1 bits (183), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 37/119 (31%), Positives = 64/119 (53%), Gaps = 7/119 (5%)
Query 1 IAQFITKWMILSILWGMGGSLSLAARSAFSEEVARFCDLPLPSQLASSDADGQTLLDFEP 60
I F+ + M+ +++W G +R S+ + + + LP + L+D+E
Sbjct 2428 IQSFVLRSMLTNLVWAFSGDGKWKSREMMSDFIRQATTISLPPNQQAC------LIDYEV 2481
Query 61 SIDDGQWHHWKSRVKQVEIDTQQVKDATLVIQTVDTLRHRDVVEGWLEEKRPFILCGPP 119
+ G W W S+V +EI++ +V A LV+ T+DT+RH ++ WL E +P +LCGPP
Sbjct 2482 QLS-GDWQPWLSKVPTMEIESHRVAAADLVVPTIDTVRHEMLLAAWLAEHKPLVLCGPP 2539
> Hs22044903
Length=2252
Score = 46.2 bits (108), Expect = 1e-05, Method: Composition-based stats.
Identities = 29/119 (24%), Positives = 58/119 (48%), Gaps = 11/119 (9%)
Query 10 ILSILWGMGGSLSLAARSAFSEEVARFCDL------PLPSQLASSDA--DGQTLL-DFEP 60
I S++W +GGS R F + R L P+P + + D + L+ D+
Sbjct 1262 IFSLIWSIGGSCDTDGRRVF-DTFIRLIILGKDDENPVPDSVGKWECPFDEKGLVYDYMY 1320
Query 61 SIDD-GQWHHWKSRVKQVEIDTQQVKDATLVIQTVDTLRHRDVVEGWLEEKRPFILCGP 118
+ + G+W HW +K + +Q+K +++ T+DT+R+ +++ + +P + GP
Sbjct 1321 ELKNKGRWVHWNELIKNTNLGDKQIKIQDIIVPTMDTIRYTFLMDLSITYAKPLLFVGP 1379
> YKR054c
Length=4092
Score = 45.4 bits (106), Expect = 2e-05, Method: Composition-based stats.
Identities = 30/118 (25%), Positives = 53/118 (44%), Gaps = 14/118 (11%)
Query 5 ITKWMILSILWGMGGSLSLAARSAFSEEVARFCDLPLPSQLASSDADGQTLLDFEPSI-- 62
IT + S+L+ + G + ++ AF + + + D Q L D+ +
Sbjct 2314 ITLLIKRSLLYALAGDSTGESQRAFIQTINTY-----------FGHDSQELSDYSTIVIA 2362
Query 63 -DDGQWHHWKSRVKQVEIDTQQVKDATLVIQTVDTLRHRDVVEGWLEEKRPFILCGPP 119
D + + S + V ++ +V +VI T+DT++H + L KR ILCGPP
Sbjct 2363 NDKLSFSSFCSEIPSVSLEAHEVMRPDIVIPTIDTIKHEKIFYDLLNSKRGIILCGPP 2420
> 7292152
Length=3868
Score = 44.3 bits (103), Expect = 6e-05, Method: Composition-based stats.
Identities = 29/118 (24%), Positives = 58/118 (49%), Gaps = 11/118 (9%)
Query 9 MILSILWGMGGSLSLAARSAFSEEVARF----CDLPLPSQLASSDADGQT---LLDFEPS 61
++ +++WG+GG L A+R F + + D P P L + T L+D+
Sbjct 1800 ILFALIWGVGGVLDTASREKFDVFLKKVWLWDTDPPPPEPLGKMEITPPTEGLLVDYVFL 1859
Query 62 IDD-GQWHHWKSRVKQVEIDTQQVKDATLVIQTVDTLRHRDVVEGWLEEKRPFILCGP 118
G W +W K+++++ + +++ TVDT R+ +++ +E K+ +L GP
Sbjct 1860 YKQRGAWRYWPDLAKRMDVEETKTG---VIVPTVDTARYIHLLKMHVEHKKRMLLVGP 1914
> SPAC1093.06c
Length=1889
Score = 40.8 bits (94), Expect = 6e-04, Method: Composition-based stats.
Identities = 28/120 (23%), Positives = 55/120 (45%), Gaps = 8/120 (6%)
Query 1 IAQFITKWMILSILWGMGGSLSLAARSAFSEEVARFCDLPLPSQLASSDADGQTLLDFEP 60
+ ++ K + + W G +R F+ + + + LP D + ++LDF+
Sbjct 103 LCNYLKKKICYILAWCCTGDTDAKSRERFTHWLMQNASVDLPE---IKDFEHVSILDFDV 159
Query 61 SIDDGQWHHWKSRVKQVEIDTQQVKDA-TLVIQTVDTLRHRDVVEGWLEEKRPFILCGPP 119
S++ W+ + + + +K A VI T+DT+R+ + + L + R I CGPP
Sbjct 160 SLETQSWYPIAGKT----LKSSALKYAGNTVIPTLDTVRYAEFLNFSLTKNRCVIFCGPP 215
> Hs15029526
Length=4490
Score = 39.3 bits (90), Expect = 0.002, Method: Composition-based stats.
Identities = 26/120 (21%), Positives = 56/120 (46%), Gaps = 12/120 (10%)
Query 3 QFITKWMILSILWGMGGSLSLAARS---AFSEEVARFCDLP-LPSQLASSDADGQTLLDF 58
+ + K + ++W +G L L +R AF + DLP +P QT+ +F
Sbjct 2344 EHLHKLFVFGLMWSLGALLELESREKLEAFLRQHESKLDLPEIPK------GSNQTMYEF 2397
Query 59 EPSIDDGQWHHWKSRVKQVEIDTQQVKD-ATLVIQTVDTLRHRDVVEGWLEEKRPFILCG 117
+ D G W HW +++ T + + +++++ VD +R +++ ++ + +L G
Sbjct 2398 YVT-DYGDWEHWNKKLQPYYYPTDSIPEYSSILVPNVDNIRTNFLIDTIAKQHKAVLLTG 2456
> Hs22057633
Length=1770
Score = 36.6 bits (83), Expect = 0.010, Method: Composition-based stats.
Identities = 32/114 (28%), Positives = 44/114 (38%), Gaps = 21/114 (18%)
Query 17 MGGSLSLAARSAFSEEVARFCDLP-------------LPSQLASSDADGQTLLDFEPSID 63
+G SL R F E + R L LP QL TL DF
Sbjct 807 LGASLLEDGRMKFDEYIKRLASLSTVDTEGVWANPGELPGQLP-------TLYDFHFDNK 859
Query 64 DGQWHHWKSRVKQVEIDTQQVKDATLVIQTVDTLRHRDVVEGWLEEKRPFILCG 117
QW W V + I + K +++ TVDT R ++E ++ K+P I G
Sbjct 860 RNQWVPWSKLVPEY-IHAPERKFINILVHTVDTTRTTWILEQMVKIKQPVIFVG 912
> Hs17864092
Length=4024
Score = 36.2 bits (82), Expect = 0.014, Method: Composition-based stats.
Identities = 26/128 (20%), Positives = 58/128 (45%), Gaps = 19/128 (14%)
Query 10 ILSILWGMGGSLSLAARSAFSEEVARFCDLPLP------------SQLASSDA------D 51
+ S++W +G S + R F++ + + P+ ++ SS A +
Sbjct 1850 LFSLIWSVGASCTDDDRLKFNKILRELMESPISDRTRNTFKLQSGTEQTSSKALTVPFPE 1909
Query 52 GQTLLDFEPSIDD-GQWHHWKSRVKQVEIDTQQVKDATLVIQTVDTLRHRDVVEGWLEEK 110
T+ D++ + G+W W ++K+ + V +++ T+DT+R+ ++E +
Sbjct 1910 KGTIYDYQFVTEGIGKWEPWIKKLKEAPPIPKDVMFNEIIVPTLDTIRYSALMELLTTHQ 1969
Query 111 RPFILCGP 118
+P I GP
Sbjct 1970 KPSIFVGP 1977
> Hs13876382
Length=4486
Score = 35.0 bits (79), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 28/115 (24%), Positives = 51/115 (44%), Gaps = 14/115 (12%)
Query 8 WMILSILWGMGGSLS----LAARSAFSEE-VARFCDLPLPSQLASSDADGQTLLDFEPSI 62
+ + + +W GG++ + R+ FS+ + F + PSQ T+ D+
Sbjct 2373 YFVFAAIWAFGGAMVQDQLVDYRAEFSKWWLTEFKTVKFPSQ--------GTIFDYYIDP 2424
Query 63 DDGQWHHWKSRVKQVEIDTQQVKDATLVIQTVDTLRHRDVVEGWLEEKRPFILCG 117
+ ++ W V Q E D + A LV T +T+R +E + +RP +L G
Sbjct 2425 ETKKFEPWSKLVPQFEFDPEMPLQACLV-HTSETIRVCYFMERLMARQRPVMLVG 2478
> Hs19115954
Length=4624
Score = 33.5 bits (75), Expect = 0.11, Method: Composition-based stats.
Identities = 23/116 (19%), Positives = 49/116 (42%), Gaps = 12/116 (10%)
Query 7 KWMILSILWGMGGSLSLAARSAFS----EEVARFCDLPLPSQLASSDADGQTLLDFEPSI 62
+ + ++LW G +L L R +LP P+ G T D+ +
Sbjct 2482 RLFVFALLWSAGAALELDGRRRLELWLRSRPTGTLELPPPA------GPGDTAFDYYVA- 2534
Query 63 DDGQWHHWKSRVKQVEIDTQQVKD-ATLVIQTVDTLRHRDVVEGWLEEKRPFILCG 117
DG W HW +R ++ + + ++++ VD +R +++ ++ + +L G
Sbjct 2535 PDGTWTHWNTRTQEYLYPSDTTPEYGSILVPNVDNVRTDFLIQTIAKQGKAVLLIG 2590
> 7301678
Length=4820
Score = 32.7 bits (73), Expect = 0.16, Method: Composition-based stats.
Identities = 25/89 (28%), Positives = 36/89 (40%), Gaps = 8/89 (8%)
Query 17 MGGSLSLAARSAFSEEVARFCDLPLPSQLASSDADG-------QTLLDFEPSIDDGQWHH 69
MG L + F E + R PL + A G TL D+ + D W
Sbjct 2957 MGACLVEKHQPVFDEYMKRISGFPLVQDTPENPASGGQFPQSKPTLYDYFWDVKDNVWKA 3016
Query 70 WKSRVKQVEIDTQQVKDATLVIQTVDTLR 98
W+ V+ D QVK + +++ TVD R
Sbjct 3017 WEWVVQPYTHDP-QVKFSEILVPTVDNTR 3044
> 7298401
Length=4010
Score = 30.4 bits (67), Expect = 0.84, Method: Composition-based stats.
Identities = 27/137 (19%), Positives = 52/137 (37%), Gaps = 36/137 (26%)
Query 10 ILSILWGMGGSLSLAARSAFS---------------------------EEVARFCDLPLP 42
+ S +W +GGSL +R F+ E +A+ P+P
Sbjct 1819 LFSCIWSLGGSLDADSREKFNIIFRALMEKTFPQSLYDTYGVPEDLYVESLAKPFIFPIP 1878
Query 43 SQLASSDADGQTLLDFEPSID-DGQWHHWKSRVKQVEIDTQQVKDATLVIQTVDTLRHRD 101
Q ++ D+ + G+W W+ V + + ++IQT +++R
Sbjct 1879 KQ--------GSVFDYRYIKEGKGKWKPWQDDVNSAPPIPRDIPVNQIIIQTNESVRIGA 1930
Query 102 VVEGWLEEKRPFILCGP 118
V++ +P +L GP
Sbjct 1931 VLDLLNRHGKPIMLVGP 1947
> Hs22067157
Length=3672
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 15/55 (27%), Positives = 27/55 (49%), Gaps = 1/55 (1%)
Query 65 GQWHHWKSRV-KQVEIDTQQVKDATLVIQTVDTLRHRDVVEGWLEEKRPFILCGP 118
G W W + K+ E K + L+I T++T R ++ +L+ + P + GP
Sbjct 1577 GHWETWTQYITKEEEKVPAGAKVSELIIPTMETARQSFFLKTYLDHEIPMLFVGP 1631
> Hs22049862
Length=891
Score = 29.6 bits (65), Expect = 1.4, Method: Composition-based stats.
Identities = 32/87 (36%), Positives = 38/87 (43%), Gaps = 14/87 (16%)
Query 25 ARSAFSEEVARFCDLP-----LPSQ-LASSDADGQTLLDF----EPSIDDGQ---WHHWK 71
AR SEE +RF L LPSQ L S D D Q LDF E DG+ + W
Sbjct 364 ARPKKSEE-SRFSHLTSLPQQLPSQQLMSKDQDEQEELDFLFDEEMEQMDGRKNTFTAWS 422
Query 72 SRVKQVEIDTQQVKDATLVIQTVDTLR 98
EID + V +V QT +R
Sbjct 423 DEESDYEIDDRDVNKILIVTQTPHYMR 449
> 7299225
Length=3508
Score = 29.6 bits (65), Expect = 1.5, Method: Composition-based stats.
Identities = 28/110 (25%), Positives = 46/110 (41%), Gaps = 7/110 (6%)
Query 10 ILSILWGMGGSLSLAARSAFSEEVAR-FCDLPLPSQLASSDADGQTLLDFEPSIDDGQWH 68
I ++ WG+GG LS + R + V F L P A T+ DF S G W
Sbjct 2475 IFALAWGLGGYLSTSDRQRMNLFVKESFPQLDYP----KGSAHENTIFDFFVS-PAGVWQ 2529
Query 69 HWKSRVKQVEIDTQQVKD-ATLVIQTVDTLRHRDVVEGWLEEKRPFILCG 117
WK+ V D ++++ VD +R ++ ++R ++ G
Sbjct 2530 SWKTLVTPYMYPELSTPDYLSILVPIVDNVRIDYLIGTIANQERAVMVIG 2579
> 7293415
Length=4081
Score = 29.3 bits (64), Expect = 1.6, Method: Composition-based stats.
Identities = 23/119 (19%), Positives = 52/119 (43%), Gaps = 14/119 (11%)
Query 3 QFITKWMILSILWGMGGSLSLAARSAFSEE----VARFCDLPLPSQLASSDADGQTLLDF 58
+ +TK ++LW + +L A + +F E+ +A+ ++ LP+ TL ++
Sbjct 1925 ELVTKIFAWAVLWAIASNLKDAEKVSFEEQWSKAIAQHPNMTLPN---------FTLWNY 1975
Query 59 EPSIDDGQWHHWKSRVKQVEIDTQQVKDATLVIQTVDTLRHRDVVEGWLEEKRPFILCG 117
++ W W + + D + + + TVDT ++ V + + P ++ G
Sbjct 1976 RIDLEKMDWGSWIDIMAKFVFDP-ETSYYDMQVPTVDTTKYGYVSDLLFKRGMPVMVTG 2033
> 7292648
Length=837
Score = 29.3 bits (64), Expect = 1.9, Method: Composition-based stats.
Identities = 18/45 (40%), Positives = 24/45 (53%), Gaps = 2/45 (4%)
Query 31 EEVARFCDLPLPSQLASSDADGQ-TLLDFEPSIDD-GQWHHWKSR 73
E F DLP P QL S+ D + +L+ + S+DD Q H SR
Sbjct 650 EHAEEFSDLPTPPQLPSTTDDNEDDVLEIQTSLDDVRQLHTPSSR 694
> SPAPB1A10.09
Length=731
Score = 29.3 bits (64), Expect = 2.1, Method: Composition-based stats.
Identities = 24/90 (26%), Positives = 42/90 (46%), Gaps = 4/90 (4%)
Query 28 AFSEEVAR-FCDLPLPSQLASSDADGQTLLDFEPSIDDGQWHHWKSRVKQVEIDTQQVKD 86
AF E+V + F + Q+ +D D +L + + + Q H + K++E D QQ D
Sbjct 32 AFREQVEKHFSKIERLHQVLGTDGDNSSLFELFTTAMNAQLHEMEQCQKKLEDDCQQRID 91
Query 87 ATLVIQTVDTLRHRDVVEGWLEEKRPFILC 116
+ + V +L+ D L+ + P I C
Sbjct 92 SIRFL--VSSLKLTDDTSS-LKIESPLIQC 118
> YDL111c
Length=265
Score = 28.1 bits (61), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 11/38 (28%), Positives = 24/38 (63%), Gaps = 0/38 (0%)
Query 68 HHWKSRVKQVEIDTQQVKDATLVIQTVDTLRHRDVVEG 105
HH ++ + QV++D +D LV++T+ +L ++ + G
Sbjct 69 HHVENELLQVDVDIAGQRDDALVVETITSLLNKVLKSG 106
> CE01861_1
Length=1827
Score = 28.1 bits (61), Expect = 3.9, Method: Composition-based stats.
Identities = 15/64 (23%), Positives = 32/64 (50%), Gaps = 6/64 (9%)
Query 47 SSDADGQTLLDFEPSIDDGQWHHWKSRVKQVEIDTQQVKDATLVIQTVDTLRHRDVVEGW 106
+SD T L+ + + D +W+H + +++ E+D L+ + DT+R W
Sbjct 1197 TSDTFQTTELENKFELLDVEWNHLEEELRRREVDLNDSMSQILIDEQFDTIRQ------W 1250
Query 107 LEEK 110
++E+
Sbjct 1251 IKER 1254
> At5g48080
Length=227
Score = 28.1 bits (61), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 16/37 (43%), Positives = 19/37 (51%), Gaps = 1/37 (2%)
Query 30 SEEVARFCDLPLPSQLASSDADGQTLLDF-EPSIDDG 65
SEE+ +FCDLPL D D L F +ID G
Sbjct 24 SEELVKFCDLPLKWNSVDYDDDSSGLSVFLRGNIDSG 60
> Hs7657351
Length=1328
Score = 27.7 bits (60), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 20/33 (60%), Gaps = 1/33 (3%)
Query 5 ITKWMILSILWGMGGSLSLAARSAFSEEVARFC 37
+ KW+I ++ + SL L A +E+VARFC
Sbjct 448 LRKWIIFRLV-SIVDSLHLEMEEALTEQVARFC 479
> CE11278
Length=401
Score = 27.3 bits (59), Expect = 6.5, Method: Composition-based stats.
Identities = 15/47 (31%), Positives = 27/47 (57%), Gaps = 1/47 (2%)
Query 30 SEEVARFCDLPLPSQLASSDADGQTLLDFEPSIDDGQWHHWKSRVKQ 76
+EE+ R ++PL S D+ GQ +L ++ SI G+ W R+++
Sbjct 107 AEELYRVFEVPLDSVTLRCDSGGQLVL-YDRSIVQGEIPFWTIRLEE 152
> Hs20538232
Length=543
Score = 27.3 bits (59), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 12/36 (33%), Positives = 23/36 (63%), Gaps = 1/36 (2%)
Query 43 SQLASSDADGQTL-LDFEPSIDDGQWHHWKSRVKQV 77
++L SS +D + L+FEP ++DG + W +++V
Sbjct 392 AELNSSPSDALLMGLNFEPEVEDGAYVKWDGFLQEV 427
> At1g31690
Length=420
Score = 26.9 bits (58), Expect = 8.2, Method: Composition-based stats.
Identities = 19/70 (27%), Positives = 31/70 (44%), Gaps = 1/70 (1%)
Query 34 ARFCDLPLPSQLASSDADGQTLLDFEPSIDDGQWHHWKSRVKQVEIDTQQVKDATLVIQT 93
A F D P Q + + FE D W H ++ V ++I T+ D +LV +
Sbjct 88 AAFMDGIFPGQDGTPTKISNVMCIFEKYAGDIMWRHTEAEVPGLKI-TEVRPDVSLVARM 146
Query 94 VDTLRHRDVV 103
V T+ + D +
Sbjct 147 VTTVGNYDYI 156
Lambda K H
0.321 0.136 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1164469306
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40