bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6636_orf1
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
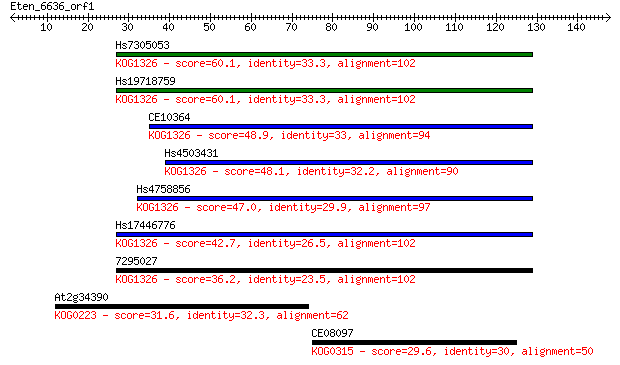
Hs7305053 60.1 1e-09
Hs19718759 60.1 1e-09
CE10364 48.9 3e-06
Hs4503431 48.1 6e-06
Hs4758856 47.0 1e-05
Hs17446776 42.7 3e-04
7295027 36.2 0.024
At2g34390 31.6 0.63
CE08097 29.6 2.0
> Hs7305053
Length=2061
Score = 60.1 bits (144), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 34/106 (32%), Positives = 50/106 (47%), Gaps = 4/106 (3%)
Query 27 IELRTLKVE-GSTVSHGTLRGFFEIMTEDYAQEHPQFTLASAEADDFQLRLVIWRVKAVP 85
+E RTL +S G L+ + ++ + P F + +A + LR++IW K V
Sbjct 1742 VETRTLHSTFQPNISQGKLQMWVDVFPKSLGPPGPPFNITPRKAKKYYLRVIIWNTKDVI 1801
Query 86 LDDSSSVSMFVRSIFT---LEENSEIVHDTDTHYNSKDGSGVFNWR 128
LD+ S + I+ + N E TD HY S DG G FNWR
Sbjct 1802 LDEKSITGEEMSDIYVKGWIPGNEENKQKTDVHYRSLDGEGNFNWR 1847
> Hs19718759
Length=2048
Score = 60.1 bits (144), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 34/106 (32%), Positives = 50/106 (47%), Gaps = 4/106 (3%)
Query 27 IELRTLKVE-GSTVSHGTLRGFFEIMTEDYAQEHPQFTLASAEADDFQLRLVIWRVKAVP 85
+E RTL +S G L+ + ++ + P F + +A + LR++IW K V
Sbjct 1729 VETRTLHSTFQPNISQGKLQMWVDVFPKSLGPPGPPFNITPRKAKKYYLRVIIWNTKDVI 1788
Query 86 LDDSSSVSMFVRSIFT---LEENSEIVHDTDTHYNSKDGSGVFNWR 128
LD+ S + I+ + N E TD HY S DG G FNWR
Sbjct 1789 LDEKSITGEEMSDIYVKGWIPGNEENKQKTDVHYRSLDGEGNFNWR 1834
> CE10364
Length=2034
Score = 48.9 bits (115), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 31/98 (31%), Positives = 49/98 (50%), Gaps = 7/98 (7%)
Query 35 EGSTVSHGTLRGFFEIMTEDYAQEHPQFTLASAEADDFQLRLVIWRVK-AVPLDDS--SS 91
+G G LR F +I +Y F +A + ++QLR+ + V+ A+P+ S
Sbjct 1665 KGGRTQKGELRMFVDIFPMEYGAIPAPFNIAPRKPINYQLRIAVMDVRGAIPVKRSFAEP 1724
Query 92 VS-MFVRSIFTLEENSEIVHDTDTHYNSKDGSGVFNWR 128
VS ++V++ H TDTH+ DG+G FNWR
Sbjct 1725 VSDLYVKAFINGMTKG---HKTDTHFRVLDGTGEFNWR 1759
> Hs4503431
Length=2080
Score = 48.1 bits (113), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 29/96 (30%), Positives = 43/96 (44%), Gaps = 9/96 (9%)
Query 39 VSHGTLRGFFEIMTEDYAQEHPQFTLASAEADDFQLRLVIWRVKAVPLDDSSSVSMFVRS 98
+ G L+ + ++ + + P F + A F LR +IW + V LDD S +
Sbjct 1778 IEQGKLQMWVDLFPKALGRPGPPFNITPRRARRFFLRCIIWNTRDVILDDLSLTGEKMSD 1837
Query 99 IFT------LEENSEIVHDTDTHYNSKDGSGVFNWR 128
I+ EE+ + TD HY S G G FNWR
Sbjct 1838 IYVKGWMIGFEEHKQ---KTDVHYRSLGGEGNFNWR 1870
> Hs4758856
Length=1230
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 29/103 (28%), Positives = 45/103 (43%), Gaps = 8/103 (7%)
Query 32 LKVEGSTVSHGTLRGFFEIMTEDYAQEHPQFTLASAEADDFQLRLVIWRVKAVPLDDS-- 89
L + + G L + ++ D ++ + ++LR++IW V L+D
Sbjct 925 LNPDKPGIEQGRLELWVDMFPMDMPAPGTPLDISPRKPKKYELRVIIWNTDEVVLEDDDF 984
Query 90 ----SSVSMFVRSIFTLEENSEIVHDTDTHYNSKDGSGVFNWR 128
S +FVR L+ E DTD HY+S G G FNWR
Sbjct 985 FTGEKSSDIFVRGW--LKGQQEDKQDTDVHYHSLTGEGNFNWR 1025
> Hs17446776
Length=802
Score = 42.7 bits (99), Expect = 3e-04, Method: Composition-based stats.
Identities = 27/109 (24%), Positives = 52/109 (47%), Gaps = 10/109 (9%)
Query 27 IELRTLKVEGST-VSHGTLRGFFEIMTEDYAQEHPQFTLASAEADDFQLRLVIWRVKAVP 85
+E RTL + G ++ + +I + PQ + + ++LR +IW+ V
Sbjct 420 VETRTLYSHSQPGIDQGKVQMWVDIFPKKLGPPGPQVNINPRKPKRYELRCIIWKTANVD 479
Query 86 LDDSS-----SVSMFVRS-IFTLEENSEIVHDTDTHYNSKDGSGVFNWR 128
L D + + ++++ ++ LE++ + TD HY+S G FNWR
Sbjct 480 LVDDNLSREKTSDIYIKGWLYGLEKD---MQKTDIHYHSLTGEADFNWR 525
> 7295027
Length=1782
Score = 36.2 bits (82), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 24/105 (22%), Positives = 46/105 (43%), Gaps = 4/105 (3%)
Query 27 IELRTL-KVEGSTVSHGTLRGFFEIMTEDYAQEHPQFTLASAEADDFQLRLVIWRVKAVP 85
+E R+L + + V G L+ + E+ + P + DF++R+V+ + +
Sbjct 1318 VETRSLCRDDFPNVEQGKLQLWIELFEANIYVPSP-IDITPVPPADFEVRVVVKNLVGIQ 1376
Query 86 LDDSSSVSMFVRSIFTLE--ENSEIVHDTDTHYNSKDGSGVFNWR 128
D + + ++ + E+ + TD HY S G FNWR
Sbjct 1377 AGDKNIFGKLMSDVYVIGWCEDEDKRQSTDIHYRSFAGDAAFNWR 1421
> At2g34390
Length=288
Score = 31.6 bits (70), Expect = 0.63, Method: Compositional matrix adjust.
Identities = 20/67 (29%), Positives = 34/67 (50%), Gaps = 5/67 (7%)
Query 12 FNPKVA-AMVEQEITPIE----LRTLKVEGSTVSHGTLRGFFEIMTEDYAQEHPQFTLAS 66
FNP V A+ + P+ T++V GST++ TLR F++ + +++H F +S
Sbjct 105 FNPAVTLALASSQRFPLNQVPAYITVQVIGSTLASATLRLLFDLNNDVCSKKHDVFLGSS 164
Query 67 AEADDFQ 73
D Q
Sbjct 165 PSGSDLQ 171
> CE08097
Length=382
Score = 29.6 bits (65), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 15/53 (28%), Positives = 28/53 (52%), Gaps = 3/53 (5%)
Query 75 RLVIWRVKAVPLDDSSSVSMFVRSIFTLEENS---EIVHDTDTHYNSKDGSGV 124
RL+ W + + +SS+ MF I +E+++ + D D HY ++G G+
Sbjct 235 RLLCWDLMTREVVESSAFDMFKSPIVPIEQDTGTYGCLRDEDKHYKMREGHGL 287
Lambda K H
0.322 0.135 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1785281974
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40