bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6644_orf1
Length=117
Score E
Sequences producing significant alignments: (Bits) Value
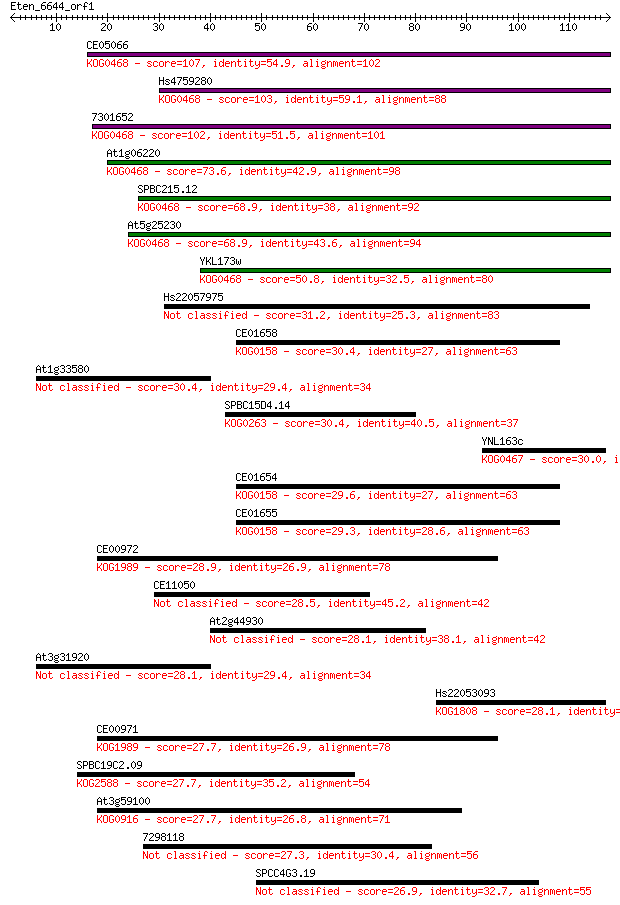
CE05066 107 4e-24
Hs4759280 103 8e-23
7301652 102 2e-22
At1g06220 73.6 9e-14
SPBC215.12 68.9 2e-12
At5g25230 68.9 2e-12
YKL173w 50.8 5e-07
Hs22057975 31.2 0.55
CE01658 30.4 0.84
At1g33580 30.4 0.91
SPBC15D4.14 30.4 0.93
YNL163c 30.0 0.96
CE01654 29.6 1.6
CE01655 29.3 1.8
CE00972 28.9 2.4
CE11050 28.5 3.2
At2g44930 28.1 3.7
At3g31920 28.1 4.3
Hs22053093 28.1 4.4
CE00971 27.7 4.9
SPBC19C2.09 27.7 5.6
At3g59100 27.7 6.0
7298118 27.3 6.4
SPCC4G3.19 26.9 9.0
> CE05066
Length=974
Score = 107 bits (268), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 56/104 (53%), Positives = 70/104 (67%), Gaps = 2/104 (1%)
Query 16 RMDEDDSQQ-QQQKPVPHELKQYYPTAAEVYPEA-ETLVQEEDLQPITEPIIAPTKVSDF 73
RM+EDD+++ Q + V HE K+YY TA EVY E ETLVQEED QP+TEPI+ P F
Sbjct 46 RMEEDDAEEIPQNQVVLHEDKKYYATALEVYGEGVETLVQEEDAQPLTEPIVKPVSKKKF 105
Query 74 DLLETQLPETTFSFDYLASLMHTPHCVRNICLLGSLHSGKTTFV 117
E LPET + +YLA LM PH +RN+ + G LH GKTTF+
Sbjct 106 QAAERFLPETVYKKEYLADLMDCPHIMRNVAIAGHLHHGKTTFL 149
> Hs4759280
Length=972
Score = 103 bits (257), Expect = 8e-23, Method: Compositional matrix adjust.
Identities = 52/89 (58%), Positives = 60/89 (67%), Gaps = 1/89 (1%)
Query 30 VPHELKQYYPTAAEVY-PEAETLVQEEDLQPITEPIIAPTKVSDFDLLETQLPETTFSFD 88
V HE K+YYPTA EVY PE ET+VQEED QP+TEPII P K F L+E LP T + D
Sbjct 58 VLHEDKKYYPTAEEVYGPEVETIVQEEDTQPLTEPIIKPVKTKKFTLMEQTLPVTVYEMD 117
Query 89 YLASLMHTPHCVRNICLLGSLHSGKTTFV 117
+LA LM +RN+ L G LH GKT FV
Sbjct 118 FLADLMDNSELIRNVTLCGHLHHGKTCFV 146
> 7301652
Length=975
Score = 102 bits (254), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 52/108 (48%), Positives = 72/108 (66%), Gaps = 7/108 (6%)
Query 17 MDEDDSQQQQQKP------VPHELKQYYPTAAEVY-PEAETLVQEEDLQPITEPIIAPTK 69
MDED+ + Q+ + V HE K+YYP+A EVY P+ ET+VQEED QP+ +P+I P K
Sbjct 41 MDEDEVEPQEDEDKEVTAVVLHEDKRYYPSAVEVYGPDVETIVQEEDAQPLDKPLIEPVK 100
Query 70 VSDFDLLETQLPETTFSFDYLASLMHTPHCVRNICLLGSLHSGKTTFV 117
F + E + ETT+ +++A LM TP +RN+ L+G LH GKTTFV
Sbjct 101 KLKFQIKEQDMQETTYDMEFMADLMDTPPLIRNVALVGHLHHGKTTFV 148
> At1g06220
Length=987
Score = 73.6 bits (179), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 42/99 (42%), Positives = 59/99 (59%), Gaps = 3/99 (3%)
Query 20 DDSQQQQQKPVPHELKQYYPTAAEVYPE-AETLVQEEDLQPITEPIIAPTKVSDFDLLET 78
+D + + Q +P + K+YYPTA EVY E ETLV +ED QP+ +PII P + F+ +
Sbjct 59 NDVEMENQIVLPED-KKYYPTAEEVYGEDVETLVMDEDEQPLEQPIIKPVRDIRFE-VGV 116
Query 79 QLPETTFSFDYLASLMHTPHCVRNICLLGSLHSGKTTFV 117
+ T S +L LM P VRN+ L+G L GKT F+
Sbjct 117 KDQATYVSTQFLIGLMSNPALVRNVALVGHLQHGKTVFM 155
> SPBC215.12
Length=983
Score = 68.9 bits (167), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 35/93 (37%), Positives = 54/93 (58%), Gaps = 1/93 (1%)
Query 26 QQKPVPHELKQYYPTAAEVY-PEAETLVQEEDLQPITEPIIAPTKVSDFDLLETQLPETT 84
Q V HE KQYYP+A EVY + +VQE+D QP+++PII P + + T +P+T
Sbjct 65 QNAVVLHEDKQYYPSAEEVYGSNVDIMVQEQDTQPLSQPIIEPIRHKRIAIETTNVPDTV 124
Query 85 FSFDYLASLMHTPHCVRNICLLGSLHSGKTTFV 117
+ ++L L+ VR+ + G LH GK+ +
Sbjct 125 YKKEFLFGLLTGTDDVRSFIVAGHLHHGKSALL 157
> At5g25230
Length=973
Score = 68.9 bits (167), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 41/96 (42%), Positives = 55/96 (57%), Gaps = 2/96 (2%)
Query 24 QQQQKPVPHELKQYYPTAAEVYPE-AETLVQEEDLQPITEPIIAPTKVSDFDLLETQLPE 82
+ Q V E K+YYP A EVY E ETLV +ED Q + +PII P + F++ +
Sbjct 46 NENQNIVLPEDKKYYPIAKEVYGEDVETLVMDEDEQSLEQPIIKPVRDIRFEVGVIKDQT 105
Query 83 TTF-SFDYLASLMHTPHCVRNICLLGSLHSGKTTFV 117
TT+ S +L LM P VRN+ L+G L GKT F+
Sbjct 106 TTYVSTLFLIGLMSNPALVRNVALVGHLQHGKTVFM 141
> YKL173w
Length=1008
Score = 50.8 bits (120), Expect = 5e-07, Method: Composition-based stats.
Identities = 26/83 (31%), Positives = 46/83 (55%), Gaps = 3/83 (3%)
Query 38 YPTAAEVYPEAETLVQEEDLQPITEPIIAPTKVSD---FDLLETQLPETTFSFDYLASLM 94
+P EV ET + P+ EP+ TK+ + F L+ +P+T ++ DY+ S+
Sbjct 68 HPYGKEVEVLMETKNTQSPQTPLVEPVTERTKLQEHTIFTQLKKNIPKTRYNRDYMLSMA 127
Query 95 HTPHCVRNICLLGSLHSGKTTFV 117
+ P + N+ ++G LHSGKT+ +
Sbjct 128 NIPERIINVGVIGPLHSGKTSLM 150
> Hs22057975
Length=347
Score = 31.2 bits (69), Expect = 0.55, Method: Compositional matrix adjust.
Identities = 21/83 (25%), Positives = 38/83 (45%), Gaps = 1/83 (1%)
Query 31 PHELKQYYPTAAEVYPEAETLVQEEDLQPITEPIIAPTKVSDFDLLETQLPETTFSFDYL 90
PH K Y A +++ E + E P + A T++ E+T SF+
Sbjct 27 PHYQKSNY-NARDIFRGCELSITSESFHPGSRKQEAATEIEKGMADGNVEVESTQSFEAT 85
Query 91 ASLMHTPHCVRNICLLGSLHSGK 113
A+ + CVR + + GS+++G+
Sbjct 86 AACVLINVCVRAVLISGSVYAGQ 108
> CE01658
Length=520
Score = 30.4 bits (67), Expect = 0.84, Method: Composition-based stats.
Identities = 17/63 (26%), Positives = 34/63 (53%), Gaps = 1/63 (1%)
Query 45 YPEAETLVQEEDLQPITEPIIAPTKVSDFDLLETQLPETTFSFDYLASLMHTPHCVRNIC 104
+PE + +QEE + +P + ++S +E + ET + LAS++H C+++
Sbjct 345 HPEIQKKLQEEVDRECPDPEVTFDQLSKLKYMECVIKETLRLYP-LASIVHNRKCMKSTT 403
Query 105 LLG 107
+LG
Sbjct 404 VLG 406
> At1g33580
Length=428
Score = 30.4 bits (67), Expect = 0.91, Method: Composition-based stats.
Identities = 10/34 (29%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 6 RSSCRRCCLVRMDEDDSQQQQQKPVPHELKQYYP 39
R +C RC R + D +++K +P ++ +Y+P
Sbjct 236 RDTCPRCSAFRWERDKHTGEEKKGIPAKVLRYFP 269
> SPBC15D4.14
Length=642
Score = 30.4 bits (67), Expect = 0.93, Method: Composition-based stats.
Identities = 15/41 (36%), Positives = 24/41 (58%), Gaps = 4/41 (9%)
Query 43 EVYPEAETLVQEEDLQPITEPIIAPTKVSDFD----LLETQ 79
E+ E E L+ E L+ ++P + PTK+ D D LLE++
Sbjct 285 ELLTEKENLINESTLESPSDPPLPPTKMKDIDYFVSLLESE 325
> YNL163c
Length=1110
Score = 30.0 bits (66), Expect = 0.96, Method: Compositional matrix adjust.
Identities = 10/24 (41%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 93 LMHTPHCVRNICLLGSLHSGKTTF 116
L + P C+RNIC++ + GKT+
Sbjct 12 LQNDPSCIRNICIVAHVDHGKTSL 35
> CE01654
Length=520
Score = 29.6 bits (65), Expect = 1.6, Method: Composition-based stats.
Identities = 17/63 (26%), Positives = 32/63 (50%), Gaps = 1/63 (1%)
Query 45 YPEAETLVQEEDLQPITEPIIAPTKVSDFDLLETQLPETTFSFDYLASLMHTPHCVRNIC 104
+PE + +QEE + +P + ++S +E + E + LASL+H C++
Sbjct 345 HPEIQKKLQEEVDRECPDPEVTFDQISKLKYMECVVKEALRMYP-LASLVHNRKCMKKTN 403
Query 105 LLG 107
+LG
Sbjct 404 VLG 406
> CE01655
Length=518
Score = 29.3 bits (64), Expect = 1.8, Method: Composition-based stats.
Identities = 18/63 (28%), Positives = 32/63 (50%), Gaps = 1/63 (1%)
Query 45 YPEAETLVQEEDLQPITEPIIAPTKVSDFDLLETQLPETTFSFDYLASLMHTPHCVRNIC 104
+PE + +QEE + +P + ++S LE + E + LASL+H C++
Sbjct 345 HPEIQKKLQEEVDRECPDPEVTFDQLSKLKYLECVVKEALRLYP-LASLVHNRKCLKTTN 403
Query 105 LLG 107
+LG
Sbjct 404 VLG 406
> CE00972
Length=690
Score = 28.9 bits (63), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 21/78 (26%), Positives = 34/78 (43%), Gaps = 4/78 (5%)
Query 18 DEDDSQQQQQKPVPHELKQYYPTAAEVYPEAETLVQEEDLQPITEPIIAPTKVSDFDLLE 77
DE D Q ++ P+ +Q + ++ P AE + P++ P+I PT + DL +
Sbjct 439 DETDGSQVRKVPIDFHHRQSFSGEEQLKPAAEA----DSAGPLSCPLIKPTDLGFTDLDK 494
Query 78 TQLPETTFSFDYLASLMH 95
LP D L H
Sbjct 495 PALPRDRAQTDGKRRLPH 512
> CE11050
Length=280
Score = 28.5 bits (62), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 26/46 (56%), Gaps = 4/46 (8%)
Query 29 PVPHELKQYYPT---AAEVYPEAETLVQEED-LQPITEPIIAPTKV 70
PVP E+ +PT ++ P AE + D L+PI EPI+ PT V
Sbjct 225 PVPPEVHTPHPTHDVKSKDEPSAEVISNTADILEPIQEPIMDPTPV 270
> At2g44930
Length=505
Score = 28.1 bits (61), Expect = 3.7, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 21/42 (50%), Gaps = 1/42 (2%)
Query 40 TAAEVYPEAETLVQEEDLQPITEPIIAPTKVSDFDLLETQLP 81
T + + E L EED+ EP + T + D LLE QLP
Sbjct 205 TTQNIKKKEEILFHEEDII-FQEPCLITTILEDLILLENQLP 245
> At3g31920
Length=725
Score = 28.1 bits (61), Expect = 4.3, Method: Composition-based stats.
Identities = 10/34 (29%), Positives = 19/34 (55%), Gaps = 0/34 (0%)
Query 6 RSSCRRCCLVRMDEDDSQQQQQKPVPHELKQYYP 39
R +C RC R + D +++K +P ++ Y+P
Sbjct 229 RDTCPRCNASRWERDKHTGEEKKGIPAKVLGYFP 262
> Hs22053093
Length=1524
Score = 28.1 bits (61), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 17/33 (51%), Gaps = 0/33 (0%)
Query 84 TFSFDYLASLMHTPHCVRNICLLGSLHSGKTTF 116
T S L + M H V++ICL+G GKT
Sbjct 43 TLSHKQLQAEMMQSHMVKDICLIGGKGCGKTVI 75
> CE00971
Length=1077
Score = 27.7 bits (60), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 21/78 (26%), Positives = 34/78 (43%), Gaps = 4/78 (5%)
Query 18 DEDDSQQQQQKPVPHELKQYYPTAAEVYPEAETLVQEEDLQPITEPIIAPTKVSDFDLLE 77
DE D Q ++ P+ +Q + ++ P AE + P++ P+I PT + DL +
Sbjct 439 DETDGSQVRKVPIDFHHRQSFSGEEQLKPAAEA----DSAGPLSCPLIKPTDLGFTDLDK 494
Query 78 TQLPETTFSFDYLASLMH 95
LP D L H
Sbjct 495 PALPRDRAQTDGKRRLPH 512
> SPBC19C2.09
Length=900
Score = 27.7 bits (60), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 19/54 (35%), Positives = 26/54 (48%), Gaps = 3/54 (5%)
Query 14 LVRMDEDDSQQQQQKPVPHELKQYYPTAAEVYPEAETLVQEEDLQPITEPIIAP 67
L D DD Q+Q K + H + P+ + YP A+ E L+ TE II P
Sbjct 153 LNEFDADDKIQRQMK-ILHSVDTIDPSTVQNYPSADMSNPEVKLK--TEEIITP 203
> At3g59100
Length=1808
Score = 27.7 bits (60), Expect = 6.0, Method: Composition-based stats.
Identities = 19/72 (26%), Positives = 37/72 (51%), Gaps = 6/72 (8%)
Query 18 DEDDSQQQQQKPVPHELKQYYPTAAEVY-PEAETLVQEEDLQPITEPIIAPTKVSDFDLL 76
+E++++ Q K P E++ YY E Y E ET + E++ + + IA +D+L
Sbjct 105 EEEETKPQLAKNDPREIQAYYQNFYEKYIKEGETSRKPEEMARLYQ--IASVL---YDVL 159
Query 77 ETQLPETTFSFD 88
+T +P ++
Sbjct 160 KTVVPSPKVDYE 171
> 7298118
Length=373
Score = 27.3 bits (59), Expect = 6.4, Method: Composition-based stats.
Identities = 17/56 (30%), Positives = 27/56 (48%), Gaps = 4/56 (7%)
Query 27 QKPVPHELKQYYPTAAEVYPEAETLVQEEDLQPITEPIIAPTKVSDFDLLETQLPE 82
+KPVP+E+K + P V E V++ P+ P+ P V +E +PE
Sbjct 266 EKPVPYEVKVHVPAPYPVIKEVPVKVEKHVPYPVKIPVEKPVHVH----IEKHVPE 317
> SPCC4G3.19
Length=759
Score = 26.9 bits (58), Expect = 9.0, Method: Composition-based stats.
Identities = 18/61 (29%), Positives = 27/61 (44%), Gaps = 6/61 (9%)
Query 49 ETLVQEEDLQPITEPIIAP------TKVSDFDLLETQLPETTFSFDYLASLMHTPHCVRN 102
E+ V+ +D +T+ I TK S F L +PE F L +++P C N
Sbjct 659 ESSVELKDRSIVTKTTIMSWSKYQGTKESLFQFLSIYIPECMLPFTKLLKSIYSPDCPTN 718
Query 103 I 103
I
Sbjct 719 I 719
Lambda K H
0.319 0.133 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1171470346
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40