bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6659_orf1
Length=95
Score E
Sequences producing significant alignments: (Bits) Value
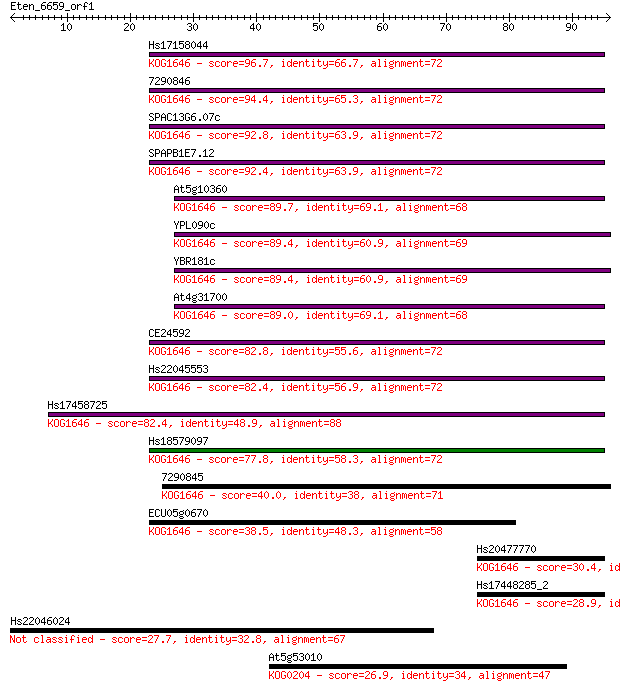
Hs17158044 96.7 9e-21
7290846 94.4 4e-20
SPAC13G6.07c 92.8 2e-19
SPAPB1E7.12 92.4 2e-19
At5g10360 89.7 1e-18
YPL090c 89.4 1e-18
YBR181c 89.4 1e-18
At4g31700 89.0 2e-18
CE24592 82.8 1e-16
Hs22045553 82.4 2e-16
Hs17458725 82.4 2e-16
Hs18579097 77.8 5e-15
7290845 40.0 0.001
ECU05g0670 38.5 0.003
Hs20477770 30.4 0.73
Hs17448285_2 28.9 2.6
Hs22046024 27.7 5.3
At5g53010 26.9 8.7
> Hs17158044
Length=249
Score = 96.7 bits (239), Expect = 9e-21, Method: Compositional matrix adjust.
Identities = 48/72 (66%), Positives = 55/72 (76%), Gaps = 0/72 (0%)
Query 23 LANHRVRLLFRTGMKCFRPRRTGERKRKSVRGAIEGPDLSVLNLVMLQKGPADIPGLTGG 82
L + RVRLL G C+RPRRTGERKRKSVRG I +LSVLNLV+++KG DIPGLT
Sbjct 68 LTHGRVRLLLSKGHSCYRPRRTGERKRKSVRGCIVDANLSVLNLVIVKKGEKDIPGLTDT 127
Query 83 EKPRRLGPKRAN 94
PRRLGPKRA+
Sbjct 128 TVPRRLGPKRAS 139
> 7290846
Length=217
Score = 94.4 bits (233), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 47/72 (65%), Positives = 55/72 (76%), Gaps = 0/72 (0%)
Query 23 LANHRVRLLFRTGMKCFRPRRTGERKRKSVRGAIEGPDLSVLNLVMLQKGPADIPGLTGG 82
L + RVRLL + G C+RPRRTGERKRKSVRG I ++SVL LV+L+KG DIPGLT
Sbjct 37 LTHGRVRLLLKKGHSCYRPRRTGERKRKSVRGCIVDANMSVLALVVLKKGEKDIPGLTDT 96
Query 83 EKPRRLGPKRAN 94
PRRLGPKRA+
Sbjct 97 TIPRRLGPKRAS 108
> SPAC13G6.07c
Length=239
Score = 92.8 bits (229), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 46/72 (63%), Positives = 54/72 (75%), Gaps = 0/72 (0%)
Query 23 LANHRVRLLFRTGMKCFRPRRTGERKRKSVRGAIEGPDLSVLNLVMLQKGPADIPGLTGG 82
L HRVRLL R G C+RPRR GERKRKSVRG I G DL+VL L ++++G DIPGLT
Sbjct 68 LLPHRVRLLLRAGHPCYRPRRDGERKRKSVRGCIVGQDLAVLALAIIKQGEQDIPGLTDV 127
Query 83 EKPRRLGPKRAN 94
P+RLGPKRA+
Sbjct 128 TVPKRLGPKRAS 139
> SPAPB1E7.12
Length=239
Score = 92.4 bits (228), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 46/72 (63%), Positives = 54/72 (75%), Gaps = 0/72 (0%)
Query 23 LANHRVRLLFRTGMKCFRPRRTGERKRKSVRGAIEGPDLSVLNLVMLQKGPADIPGLTGG 82
L HRVRLL R G C+RPRR GERKRKSVRG I G DL+VL L ++++G DIPGLT
Sbjct 68 LLPHRVRLLLRAGHPCYRPRRDGERKRKSVRGCIVGQDLAVLALAIVKQGEQDIPGLTDV 127
Query 83 EKPRRLGPKRAN 94
P+RLGPKRA+
Sbjct 128 TVPKRLGPKRAS 139
> At5g10360
Length=249
Score = 89.7 bits (221), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 47/70 (67%), Positives = 54/70 (77%), Gaps = 2/70 (2%)
Query 27 RVRLLFRTGMKCFRP--RRTGERKRKSVRGAIEGPDLSVLNLVMLQKGPADIPGLTGGEK 84
RVRLL G CFR RRTGER+RKSVRG I PDLSVLNLV+++KG +D+PGLT EK
Sbjct 72 RVRLLLHRGTPCFRGHGRRTGERRRKSVRGCIVSPDLSVLNLVIVKKGVSDLPGLTDTEK 131
Query 85 PRRLGPKRAN 94
PR GPKRA+
Sbjct 132 PRMRGPKRAS 141
> YPL090c
Length=236
Score = 89.4 bits (220), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 42/69 (60%), Positives = 54/69 (78%), Gaps = 0/69 (0%)
Query 27 RVRLLFRTGMKCFRPRRTGERKRKSVRGAIEGPDLSVLNLVMLQKGPADIPGLTGGEKPR 86
R++LL + C+RPRR GERKRKSVRGAI GPDL+VL LV+++KG ++ GLT P+
Sbjct 72 RIKLLLTKNVSCYRPRRDGERKRKSVRGAIVGPDLAVLALVIVKKGEQELEGLTDTTVPK 131
Query 87 RLGPKRANH 95
RLGPKRAN+
Sbjct 132 RLGPKRANN 140
> YBR181c
Length=236
Score = 89.4 bits (220), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 42/69 (60%), Positives = 54/69 (78%), Gaps = 0/69 (0%)
Query 27 RVRLLFRTGMKCFRPRRTGERKRKSVRGAIEGPDLSVLNLVMLQKGPADIPGLTGGEKPR 86
R++LL + C+RPRR GERKRKSVRGAI GPDL+VL LV+++KG ++ GLT P+
Sbjct 72 RIKLLLTKNVSCYRPRRDGERKRKSVRGAIVGPDLAVLALVIVKKGEQELEGLTDTTVPK 131
Query 87 RLGPKRANH 95
RLGPKRAN+
Sbjct 132 RLGPKRANN 140
> At4g31700
Length=250
Score = 89.0 bits (219), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 47/70 (67%), Positives = 53/70 (75%), Gaps = 2/70 (2%)
Query 27 RVRLLFRTGMKCFRP--RRTGERKRKSVRGAIEGPDLSVLNLVMLQKGPADIPGLTGGEK 84
RVRLL G CFR RRTGER+RKSVRG I PDLSVLNLV+++KG D+PGLT EK
Sbjct 72 RVRLLLHRGTPCFRGHGRRTGERRRKSVRGCIVSPDLSVLNLVIVKKGENDLPGLTDTEK 131
Query 85 PRRLGPKRAN 94
PR GPKRA+
Sbjct 132 PRMRGPKRAS 141
> CE24592
Length=246
Score = 82.8 bits (203), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 40/72 (55%), Positives = 52/72 (72%), Gaps = 0/72 (0%)
Query 23 LANHRVRLLFRTGMKCFRPRRTGERKRKSVRGAIEGPDLSVLNLVMLQKGPADIPGLTGG 82
L N RVRLL + G C+R R+ GERKRKSVRG I ++S L+LV+++KG +I GLT
Sbjct 68 LTNGRVRLLLKKGQSCYRERKNGERKRKSVRGCIVDANMSALSLVIVKKGDGEIEGLTDS 127
Query 83 EKPRRLGPKRAN 94
PR+LGPKRA+
Sbjct 128 VLPRKLGPKRAS 139
> Hs22045553
Length=90
Score = 82.4 bits (202), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 41/72 (56%), Positives = 49/72 (68%), Gaps = 0/72 (0%)
Query 23 LANHRVRLLFRTGMKCFRPRRTGERKRKSVRGAIEGPDLSVLNLVMLQKGPADIPGLTGG 82
L + V LL G C+RPRRTGERK KSV G I +LS+LNLV+++KG DIPGLT
Sbjct 6 LTHGHVHLLLSKGHSCYRPRRTGERKGKSVHGCIVVANLSILNLVIVKKGEKDIPGLTDT 65
Query 83 EKPRRLGPKRAN 94
P LGPKRA+
Sbjct 66 MVPHHLGPKRAS 77
> Hs17458725
Length=248
Score = 82.4 bits (202), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 43/89 (48%), Positives = 56/89 (62%), Gaps = 1/89 (1%)
Query 7 ELGANAAAAGAEGEEDLANH-RVRLLFRTGMKCFRPRRTGERKRKSVRGAIEGPDLSVLN 65
E+ A+A +G + H V LL C+RPRRTG+RK+KS RG I +LS+LN
Sbjct 34 EVAADALGKEWKGYVGVLTHGHVHLLLNKRHACYRPRRTGKRKQKSARGCIVDANLSILN 93
Query 66 LVMLQKGPADIPGLTGGEKPRRLGPKRAN 94
LV+++KG DIPGLT P LGPKRA+
Sbjct 94 LVIVKKGEKDIPGLTDTMVPCHLGPKRAS 122
> Hs18579097
Length=240
Score = 77.8 bits (190), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 42/75 (56%), Positives = 51/75 (68%), Gaps = 3/75 (4%)
Query 23 LANHRVRLLFRTGMKCFRPRRTGERKRKSVRGAIEGPDLSVLNLVM---LQKGPADIPGL 79
L + RV LL G C+RPRRTGERKRKSVRG I +LS+LNL++ +K DIPGL
Sbjct 68 LTHGRVHLLLSKGHSCYRPRRTGERKRKSVRGCIVDANLSILNLIIVKKKKKVKKDIPGL 127
Query 80 TGGEKPRRLGPKRAN 94
T P RLGPK+A+
Sbjct 128 TDTMVPCRLGPKKAS 142
> 7290845
Length=135
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 27/71 (38%), Positives = 38/71 (53%), Gaps = 11/71 (15%)
Query 25 NHRVRLLFRTGMKCFRPRRTGERKRKSVRGAIEGPDLSVLNLVMLQKGPADIPGLTGGEK 84
N R+RLL + CF PR RK K+VR ++S L LV+L+K P+
Sbjct 5 NRRLRLLKKIH-SCFHPRCNKVRKCKTVRKYTVEANVSALTLVVLKKNPS---------- 53
Query 85 PRRLGPKRANH 95
P RLGP R+++
Sbjct 54 PCRLGPVRSSN 64
> ECU05g0670
Length=203
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 28/58 (48%), Positives = 37/58 (63%), Gaps = 0/58 (0%)
Query 23 LANHRVRLLFRTGMKCFRPRRTGERKRKSVRGAIEGPDLSVLNLVMLQKGPADIPGLT 80
L RVR L G +R RR G R+RKSVRG+I ++ VLNL++L+ G +I GLT
Sbjct 53 LTKKRVRPLLSKGDAGYRCRRKGVRRRKSVRGSIVSEEICVLNLIILRPGEKEIDGLT 110
> Hs20477770
Length=237
Score = 30.4 bits (67), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 14/20 (70%), Positives = 15/20 (75%), Gaps = 0/20 (0%)
Query 75 DIPGLTGGEKPRRLGPKRAN 94
DIPGLTG P LGPKRA+
Sbjct 142 DIPGLTGTTVPCCLGPKRAS 161
> Hs17448285_2
Length=155
Score = 28.9 bits (63), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 12/20 (60%), Positives = 14/20 (70%), Gaps = 0/20 (0%)
Query 75 DIPGLTGGEKPRRLGPKRAN 94
+IPGLT P LGPKRA+
Sbjct 73 NIPGLTDTTMPHHLGPKRAS 92
> Hs22046024
Length=382
Score = 27.7 bits (60), Expect = 5.3, Method: Composition-based stats.
Identities = 22/73 (30%), Positives = 32/73 (43%), Gaps = 6/73 (8%)
Query 1 PKYAEMELGANAAAAGAEGEEDLANHRVRLLFRTGMKC--FRPRRTGERKRKSVRGAI-- 56
P YA + A + EDL + L ++ KC F + T + K K +G +
Sbjct 149 PTYAGSKSAAERLKRALKATEDLREEAAQALAQSDRKCHEFNRQVTVQWKEKDFQGMLKC 208
Query 57 --EGPDLSVLNLV 67
EG L +LNLV
Sbjct 209 HKEGEALLILNLV 221
> At5g53010
Length=1085
Score = 26.9 bits (58), Expect = 8.7, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 20/47 (42%), Gaps = 0/47 (0%)
Query 42 RRTGERKRKSVRGAIEGPDLSVLNLVMLQKGPADIPGLTGGEKPRRL 88
R E+KRKS G IE L L P ++ L +PR L
Sbjct 653 RAINEQKRKSFEGTIENMSKEGLRCAALAYQPCELGSLPTITEPRNL 699
Lambda K H
0.317 0.137 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1161385214
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40