bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6726_orf2
Length=152
Score E
Sequences producing significant alignments: (Bits) Value
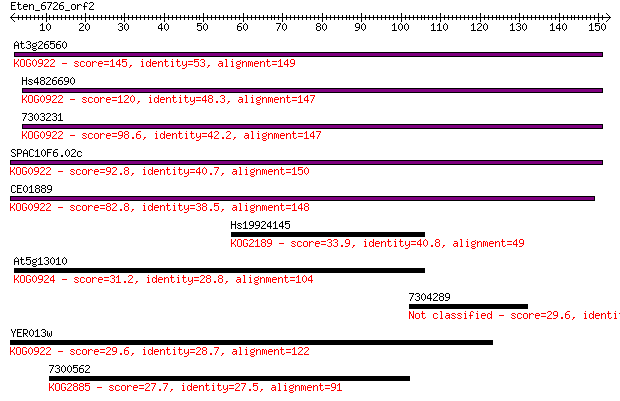
At3g26560 145 4e-35
Hs4826690 120 1e-27
7303231 98.6 4e-21
SPAC10F6.02c 92.8 2e-19
CE01889 82.8 2e-16
Hs19924145 33.9 0.13
At5g13010 31.2 0.77
7304289 29.6 2.1
YER013w 29.6 2.3
7300562 27.7 9.7
> At3g26560
Length=1168
Score = 145 bits (365), Expect = 4e-35, Method: Compositional matrix adjust.
Identities = 79/150 (52%), Positives = 112/150 (74%), Gaps = 2/150 (1%)
Query 2 KLMSDYDKWEAQQLQRSGLVSSKENPYYDEEAGGIL-PAQEAEEDVEIEVVDEEPLFIRG 60
K MS ++WEA+QL SG++ E P YDE+ G+L + AEE++EIE+ ++EP F++G
Sbjct 343 KKMSSPERWEAKQLIASGVLRVDEFPMYDEDGDGMLYQEEGAEEELEIEMNEDEPAFLQG 402
Query 61 QTTRAGMQLSPVKIVANPDGSLARAAATAQTLAKERRDVRQAQEAAILDSIPKDMSRPWE 120
QT R + +SPVKI NP+GSL+RAAA L KERR++R+ Q+ +LDSIPKD++RPWE
Sbjct 403 QT-RYSVDMSPVKIFKNPEGSLSRAAALQSALTKERREMREQQQRTMLDSIPKDLNRPWE 461
Query 121 DPNPQPGERTIAQALQGLGQTGYEMPEWKR 150
DP P+ GER +AQ L+G+G + Y+MPEWK+
Sbjct 462 DPMPETGERHLAQELRGVGLSAYDMPEWKK 491
> Hs4826690
Length=1220
Score = 120 bits (300), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 71/149 (47%), Positives = 103/149 (69%), Gaps = 4/149 (2%)
Query 4 MSDYDKWEAQQLQRSGLVSSKENPYYDEEAGGILP--AQEAEEDVEIEVVDEEPLFIRGQ 61
+SD +KWE +Q+ + ++S +E P +DEE G ILP E +ED+EIE+V+EEP F+RG
Sbjct 394 ISDPEKWEIKQMIAANVLSKEEFPDFDEETG-ILPKVDDEEDEDLEIELVEEEPPFLRGH 452
Query 62 TTRAGMQLSPVKIVANPDGSLARAAATAQTLAKERRDVRQAQEAAILDSIPKDMSRPWED 121
T + M +SP+KIV NPDGSL++AA LAKERR+++QAQ A +DSIP +++ W D
Sbjct 453 T-KQSMDMSPIKIVKNPDGSLSQAAMMQSALAKERRELKQAQREAEMDSIPMGLNKHWVD 511
Query 122 PNPQPGERTIAQALQGLGQTGYEMPEWKR 150
P P R IA ++G+G ++PEWK+
Sbjct 512 PLPDAEGRQIAANMRGIGMMPNDIPEWKK 540
> 7303231
Length=1242
Score = 98.6 bits (244), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 62/148 (41%), Positives = 96/148 (64%), Gaps = 3/148 (2%)
Query 4 MSDYDKWEAQQLQRSGLVSSKENPYYDEEAGGILPAQEAEE-DVEIEVVDEEPLFIRGQT 62
+S ++WE +Q+ SG++ E P +DEE G +LP E +E D+EIE+V+EEP F+ G
Sbjct 416 ISSPERWEIKQMISSGVLDRSEMPDFDEETG-LLPKDEDDEADIEIEIVEEEPPFLSGHG 474
Query 63 TRAGMQLSPVKIVANPDGSLARAAATAQTLAKERRDVRQAQEAAILDSIPKDMSRPWEDP 122
RA LSPV+IV NPDGSLA+AA L+KERR+ + Q ++++P +++ W DP
Sbjct 475 -RALHDLSPVRIVKNPDGSLAQAAMMQSALSKERREQKMLQREQEIEAMPTSLNKNWIDP 533
Query 123 NPQPGERTIAQALQGLGQTGYEMPEWKR 150
P+ R++A ++G+ E+PEWK+
Sbjct 534 LPEDESRSLAANMRGMAAAPPEVPEWKK 561
> SPAC10F6.02c
Length=1168
Score = 92.8 bits (229), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 61/159 (38%), Positives = 92/159 (57%), Gaps = 12/159 (7%)
Query 1 RKLMSDYDKWEAQQLQRSGLVSSKENP-----YYDEEAGGILPAQEAEEDVEIEVVDEEP 55
RK ++ + WE QQL SG +S+ + P + A I P E +EDVEIE+ +EEP
Sbjct 331 RKRLTSPEIWELQQLAASGAISATDIPELNDGFNTNNAAEINP--EDDEDVEIELREEEP 388
Query 56 LFIRGQTTRAGMQLSPVKIVANPDGSLARAAATAQTLAKERRDVRQ--AQEAAILDSIPK 113
F+ GQT + ++LSP+K+V PDGSL+RAA Q LA +RR++RQ A+ + + +
Sbjct 389 GFLAGQT-KVSLKLSPIKVVKAPDGSLSRAAMQGQILANDRREIRQKEAKLKSEQEMEKQ 447
Query 114 DMSRPWEDPNPQPGERTIAQALQGLG--QTGYEMPEWKR 150
D+S W+D P +R AQ ++ Q E P W++
Sbjct 448 DLSLSWQDTMSNPQDRKFAQDVRDSAARQLTSETPSWRQ 486
> CE01889
Length=1200
Score = 82.8 bits (203), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 57/155 (36%), Positives = 92/155 (59%), Gaps = 10/155 (6%)
Query 1 RKLMSDYDKWEAQQLQRSGLVSSKENPYYDEEAGGILPAQEAE---EDVEIEVVDEEPLF 57
R +S ++WE +Q+Q +G++++ + P +DEE G +L + E ED+EIE+V++EP F
Sbjct 362 RVRISTPERWELRQMQGAGVLTATDMPDFDEEMG-VLRNYDDESDGEDIEIELVEDEPDF 420
Query 58 IRGQTTRAGMQLSPVKIVANPDGSLARAAATAQTLAKERRDVR-QAQEAAILDSIPKDMS 116
+RG + G ++ PVK+V NPDGSLA+AA L+KER++ + QAQ +D+ K S
Sbjct 421 LRGYG-KGGAEIEPVKVVKNPDGSLAQAALMQGALSKERKETKIQAQRERDMDT-QKGFS 478
Query 117 RPWEDPNPQPGERTIAQAL---QGLGQTGYEMPEW 148
+P G ++ A + + EMPEW
Sbjct 479 SNARILDPMSGNQSTAWSADESKDRNNKMKEMPEW 513
> Hs19924145
Length=830
Score = 33.9 bits (76), Expect = 0.13, Method: Composition-based stats.
Identities = 20/51 (39%), Positives = 28/51 (54%), Gaps = 2/51 (3%)
Query 57 FIRGQTTRAGMQLSPVK--IVANPDGSLARAAATAQTLAKERRDVRQAQEA 105
F++ + RAG+ L P K + A P L R + LA+E RDVR Q+A
Sbjct 67 FLQEEVRRAGLVLPPPKGRLPAPPPRDLLRIQEETERLAQELRDVRGNQQA 117
> At5g13010
Length=1226
Score = 31.2 bits (69), Expect = 0.77, Method: Compositional matrix adjust.
Identities = 30/105 (28%), Positives = 46/105 (43%), Gaps = 11/105 (10%)
Query 2 KLMSDYDKWEAQQLQRSGLVSSKE-NPYYDEEAGGILPAQEAEEDVEIEVVDEEPLFIRG 60
+L +D +WE +QL RSG V E +D E E + V D +P F+ G
Sbjct 367 QLNADNAQWEDRQLLRSGAVRGTEVQTEFD---------SEEERKAILLVHDTKPPFLDG 417
Query 61 QTTRAGMQLSPVKIVANPDGSLARAAATAQTLAKERRDVRQAQEA 105
+ Q PV V +P +A + L KE R+ + A ++
Sbjct 418 RVVYTK-QAEPVMPVKDPTSDMAIISRKGSGLVKEIREKQSANKS 461
> 7304289
Length=941
Score = 29.6 bits (65), Expect = 2.1, Method: Composition-based stats.
Identities = 15/33 (45%), Positives = 21/33 (63%), Gaps = 3/33 (9%)
Query 102 AQEAAILDSIPKDMSRP---WEDPNPQPGERTI 131
AQ ++ S PKDMS+ WE+P+P P R+I
Sbjct 296 AQPVSVGVSGPKDMSKQISGWEEPSPPPQRRSI 328
> YER013w
Length=1145
Score = 29.6 bits (65), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 35/149 (23%), Positives = 61/149 (40%), Gaps = 29/149 (19%)
Query 1 RKLMSDYDKWEAQQLQRSGLVSSKENPYYDEEA-------------GGI----------- 36
R+ ++ ++WE +QL SG S + P +E G I
Sbjct 288 RRALTSPERWEIRQLIASGAASIDDYPELKDEIPINTSYLTAKRDDGSIVNGNTEKVDSK 347
Query 37 LPAQEAEEDVEIEV---VDEEPLFIRGQTTRAGMQLSPVKIVANPDGSLARAAATAQTLA 93
L Q+ +E EI+V D+ P F++ Q + + KI P G + R+A
Sbjct 348 LEEQQRDETDEIDVELNTDDGPKFLKDQQVKGAKKYEMPKITKVPRGFMNRSAINGSNAI 407
Query 94 KERRDVRQAQEAAILDSIPKDMSRPWEDP 122
++ R+ + ++ I I K S ++DP
Sbjct 408 RDHREEKLRKKREIEQQIRKQQS--FDDP 434
> 7300562
Length=324
Score = 27.7 bits (60), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 25/103 (24%), Positives = 50/103 (48%), Gaps = 16/103 (15%)
Query 11 EAQQLQRSG-----LVSS-----KENPYYDEEAGGILPAQEAEEDV--EIEVVDEEPLFI 58
E+++L+RS L+SS EN + + G++ +++ ED +I+ V +P++
Sbjct 119 ESKKLKRSKGVQKLLLSSAKNLKNENVKHQKLKNGVVKSEQETEDSKEQIQPVKVQPVY- 177
Query 59 RGQTTRAGMQLSPVKIVANPDGSLARAAATAQTLAKERRDVRQ 101
A + S V ANP G ++ + + K+ RD ++
Sbjct 178 ---NQEAKIVYSKVDFAANPGGKAKKSHQNPKEILKKLRDTKK 217
Lambda K H
0.310 0.129 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1925034518
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40