bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6765_orf1
Length=177
Score E
Sequences producing significant alignments: (Bits) Value
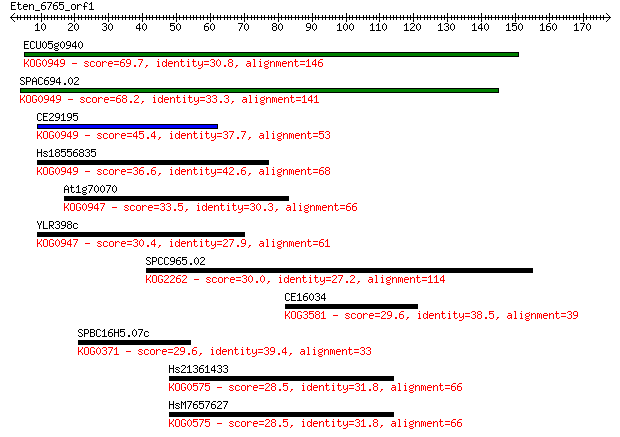
ECU05g0940 69.7 3e-12
SPAC694.02 68.2 7e-12
CE29195 45.4 5e-05
Hs18556835 36.6 0.025
At1g70070 33.5 0.24
YLR398c 30.4 1.8
SPCC965.02 30.0 2.7
CE16034 29.6 3.0
SPBC16H5.07c 29.6 3.7
Hs21361433 28.5 7.1
HsM7657627 28.5 7.3
> ECU05g0940
Length=1337
Score = 69.7 bits (169), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 45/153 (29%), Positives = 82/153 (53%), Gaps = 9/153 (5%)
Query 5 GPKVERIIQMLPCPFLAMSATVGNPDSFVGWLRRCA-PRG-DVRLVCHKERFSDLHMHLY 62
G +ERI+ + PCP L +SAT+GN + F GW+++ +G D LV + ER+ +L ++Y
Sbjct 651 GIPIERIVHLSPCPMLVLSATLGNIEGFYGWMKKIEREKGRDCELVSYGERYCELKPYVY 710
Query 63 LQD---KVLPFNPVAALHFNKVLGEGLAEDFYLPPVDSLNLYLALSRILKTS--NLFSFL 117
K++P N + A ++ + G D P + L+LY ++ +L+ +L S +
Sbjct 711 SHSFSPKMVPINNLFAYSYSHIKRFGFGNDISFLPEELLHLYDSICAVLRKDQEHLISKI 770
Query 118 FNPSFYFQGTPAITKKQYSFYWQTMRAHFVYMV 150
P +F+ + +TKK+ Y + + F M+
Sbjct 771 -TPENFFR-SNIVTKKEVGEYGRHLLETFQGMI 801
> SPAC694.02
Length=1717
Score = 68.2 bits (165), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 47/156 (30%), Positives = 75/156 (48%), Gaps = 19/156 (12%)
Query 4 EGPKVERIIQMLPCPFLAMSATVGNPDSFVGWLR--RCAPRGDVRLVCHKERFSDLHMHL 61
+G E+++ + PCP +A+SATVGNP F WL + A + L+ H+ R+SDL +
Sbjct 874 DGLVEEQLLLLAPCPIVALSATVGNPQEFQVWLSALQRAHSYPIHLIVHEHRYSDLRKFV 933
Query 62 Y--------LQD-----KVLPFNPVAALHFNKVLGEGLAEDFYLPPVDSLNLYLALSRIL 108
Y LQ+ ++ +P AA+ F+ +G + P D L LY A+ +
Sbjct 934 YGGAKEFNGLQEEGDLGQIAFIHPAAAMSFS----DGSTTNLAFEPRDCLQLYYAMKLVS 989
Query 109 KTSNLFSFLFNPSFYFQGTPAITKKQYSFYWQTMRA 144
K S +P YF+ P I K Y + ++A
Sbjct 990 KGDFKISRKLDPDRYFEDIPFIKKVDAVGYEKKIKA 1025
> CE29195
Length=1714
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 20/58 (34%), Positives = 36/58 (62%), Gaps = 5/58 (8%)
Query 9 ERIIQMLPCPFLAMSATVGNPDSFVGWLR-----RCAPRGDVRLVCHKERFSDLHMHL 61
E+++ ++ CPFLA+SAT+GN + WL + A + V L+ + ER+S+L + +
Sbjct 928 EQLLLLIQCPFLALSATIGNANKLHEWLNSSEQAKSAGKRKVELINYGERYSELELSI 985
> Hs18556835
Length=556
Score = 36.6 bits (83), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 29/106 (27%), Positives = 44/106 (41%), Gaps = 38/106 (35%)
Query 9 ERIIQMLPCPFLAMSATVGNPDSFVGWLR---------------RCAPRG---------- 43
E ++ ++ CPFL +SAT+ NP+ WL+ +C
Sbjct 447 ELLLVIIRCPFLVLSATINNPNLLTKWLQSVKQYWKQADKIMEEKCISEKQADKCLNFLQ 506
Query 44 ---------DVRLVCHKERFSDLHMHL--YLQDKVL--PFNPVAAL 76
+VRLV + ER++DL H+ D V F+P AAL
Sbjct 507 DHSYKNQSYEVRLVLYGERYNDLEKHICSVKHDDVYFDHFHPCAAL 552
> At1g70070
Length=1171
Score = 33.5 bits (75), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 20/70 (28%), Positives = 29/70 (41%), Gaps = 6/70 (8%)
Query 17 CP----FLAMSATVGNPDSFVGWLRRCAPRGDVRLVCHKERFSDLHMHLYLQDKVLPFNP 72
CP + +SATV NPD GW+ G LV R L + + +LP
Sbjct 296 CPKEVQLICLSATVANPDELAGWIGEI--HGKTELVTSTRRPVPLTWYFSTKHSLLPLLD 353
Query 73 VAALHFNKVL 82
++ N+ L
Sbjct 354 EKGINVNRKL 363
> YLR398c
Length=1287
Score = 30.4 bits (67), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 17/63 (26%), Positives = 35/63 (55%), Gaps = 3/63 (4%)
Query 9 ERIIQMLP--CPFLAMSATVGNPDSFVGWLRRCAPRGDVRLVCHKERFSDLHMHLYLQDK 66
E +I MLP F+ +SATV N F W+ R + ++ ++ +R L ++++ + +
Sbjct 459 EEVIIMLPQHVKFILLSATVPNTYEFANWIGRTKQK-NIYVISTPKRPVPLEINIWAKKE 517
Query 67 VLP 69
++P
Sbjct 518 LIP 520
> SPCC965.02
Length=791
Score = 30.0 bits (66), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 31/130 (23%), Positives = 56/130 (43%), Gaps = 17/130 (13%)
Query 41 PRGDVRLVCHKERFSDLHMHLYLQDKVLPF-----NPVAALHFNKVLGEGLAE------D 89
P ++ +CH ++SD+H L + K + + + + +L EG +
Sbjct 444 PTDVIQPICHVFKYSDIHTKLLKKYKRVHWGFYLASIIVSLGLGFAFTEGWHDIQIRSYG 503
Query 90 FYLPPVDSLNLYLALSRILKTSNLFS-----FLFNPSFYFQGTPAITKKQYSFYWQTMRA 144
F + V LY+ LS I S+ F +F+F G P YSF + T++
Sbjct 504 FVVSMVIGAALYIPLSLIESRSSFTISMQAFFEIVAAFWFNGQPMALLYFYSFGFGTLQ- 562
Query 145 HFVYMVRESK 154
H ++M + +K
Sbjct 563 HAMHMTQSAK 572
> CE16034
Length=372
Score = 29.6 bits (65), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 22/39 (56%), Gaps = 2/39 (5%)
Query 82 LGEGLAEDFYLPPVDSLNLYLALSRILKTSNLFSFLFNP 120
LGEG ++F PP+D Y+ +RI +L + FNP
Sbjct 117 LGEGKTKEF--PPLDPKGKYIKSTRIRCGRSLNGYQFNP 153
> SPBC16H5.07c
Length=322
Score = 29.6 bits (65), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 13/35 (37%), Positives = 21/35 (60%), Gaps = 2/35 (5%)
Query 21 AMSATVGNPDSFVGWLRRCAP--RGDVRLVCHKER 53
A AT+G+ D ++ L++C P DV ++C K R
Sbjct 15 ANDATLGDVDRWIEQLKKCEPLSEADVEMLCDKAR 49
> Hs21361433
Length=970
Score = 28.5 bits (62), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 21/66 (31%), Positives = 33/66 (50%), Gaps = 8/66 (12%)
Query 48 VCHKERFSDLHMHLYLQDKVLPFNPVAALHFNKVLGEGLAEDFYLPPVDSLNLYLALSRI 107
+CH + M+ YL+++V PF+ A HF + G+ YL L+ L LS +
Sbjct 91 MCH-----NGEMNRYLKNRVKPFSENEARHFMHQIITGM---LYLHSHGILHRDLTLSNL 142
Query 108 LKTSNL 113
L T N+
Sbjct 143 LLTRNM 148
> HsM7657627
Length=970
Score = 28.5 bits (62), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 21/66 (31%), Positives = 33/66 (50%), Gaps = 8/66 (12%)
Query 48 VCHKERFSDLHMHLYLQDKVLPFNPVAALHFNKVLGEGLAEDFYLPPVDSLNLYLALSRI 107
+CH + M+ YL+++V PF+ A HF + G+ YL L+ L LS +
Sbjct 91 MCH-----NGEMNRYLKNRVKPFSENEARHFMHQIITGM---LYLHSHGILHRDLTLSNL 142
Query 108 LKTSNL 113
L T N+
Sbjct 143 LLTRNM 148
Lambda K H
0.325 0.139 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2743263016
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40