bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_7183_orf1
Length=91
Score E
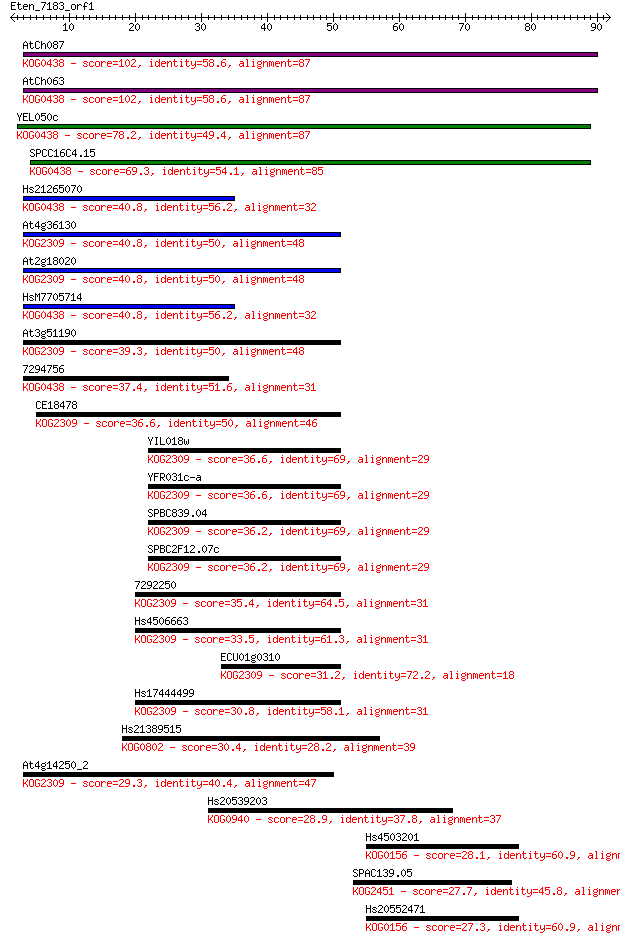
Sequences producing significant alignments: (Bits) Value
AtCh087 102 1e-22
AtCh063 102 1e-22
YEL050c 78.2 3e-15
SPCC16C4.15 69.3 1e-12
Hs21265070 40.8 5e-04
At4g36130 40.8 6e-04
At2g18020 40.8 6e-04
HsM7705714 40.8 7e-04
At3g51190 39.3 0.002
7294756 37.4 0.007
CE18478 36.6 0.010
YIL018w 36.6 0.012
YFR031c-a 36.6 0.012
SPBC839.04 36.2 0.017
SPBC2F12.07c 36.2 0.017
7292250 35.4 0.024
Hs4506663 33.5 0.10
ECU01g0310 31.2 0.45
Hs17444499 30.8 0.72
Hs21389515 30.4 0.87
At4g14250_2 29.3 1.7
Hs20539203 28.9 2.5
Hs4503201 28.1 4.6
SPAC139.05 27.7 5.4
Hs20552471 27.3 6.1
> AtCh087
Length=274
Score = 102 bits (255), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 51/88 (57%), Positives = 66/88 (75%), Gaps = 1/88 (1%)
Query 3 CMATIGEVGNSEHSLVQIGKAGRNRHMGRRPHVRGSVMNPVDHPHGGGEGKNPVGRKAPL 62
C AT+G+VGN + +G+AG +G+RP VRG VMNPVDHPHGGGEG+ P+GRK P+
Sbjct 186 CSATVGQVGNVGVNQKSLGRAGSKCWLGKRPVVRGVVMNPVDHPHGGGEGRAPIGRKKPV 245
Query 63 TPWGKPALGVKTRG-KKTTDKFIVRRRN 89
TPWG PALG +TR KK ++ I+RRR+
Sbjct 246 TPWGYPALGRRTRKRKKYSETLILRRRS 273
> AtCh063
Length=274
Score = 102 bits (255), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 51/88 (57%), Positives = 66/88 (75%), Gaps = 1/88 (1%)
Query 3 CMATIGEVGNSEHSLVQIGKAGRNRHMGRRPHVRGSVMNPVDHPHGGGEGKNPVGRKAPL 62
C AT+G+VGN + +G+AG +G+RP VRG VMNPVDHPHGGGEG+ P+GRK P+
Sbjct 186 CSATVGQVGNVGVNQKSLGRAGSKCWLGKRPVVRGVVMNPVDHPHGGGEGRAPIGRKKPV 245
Query 63 TPWGKPALGVKTRG-KKTTDKFIVRRRN 89
TPWG PALG +TR KK ++ I+RRR+
Sbjct 246 TPWGYPALGRRTRKRKKYSETLILRRRS 273
> YEL050c
Length=393
Score = 78.2 bits (191), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 43/87 (49%), Positives = 55/87 (63%), Gaps = 1/87 (1%)
Query 2 ECMATIGEVGNSEHSLVQIGKAGRNRHMGRRPHVRGSVMNPVDHPHGGGEGKNPVGRKAP 61
E +ATIG V N +H +GKAGR+R +G RP VRG MN DHPHGGG GK+ K
Sbjct 300 EAVATIGVVSNIDHQNRSLGKAGRSRWLGIRPTVRGVAMNKCDHPHGGGRGKSK-SNKLS 358
Query 62 LTPWGKPALGVKTRGKKTTDKFIVRRR 88
++PWG+ A G KTR K ++ V+ R
Sbjct 359 MSPWGQLAKGYKTRRGKNQNRMKVKDR 385
> SPCC16C4.15
Length=318
Score = 69.3 bits (168), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 46/86 (53%), Positives = 55/86 (63%), Gaps = 2/86 (2%)
Query 4 MATIGEVGNSEHSLVQIGKAGRNRHMGRRPHVRGSVMNPVDHPHGGGEGKNPVGRKAPLT 63
ATIG V N Q+GKAGR+R +G RP VRG+ MNP DHPHGGG GK+ +G K +
Sbjct 227 FATIGVVSNIYWQHRQLGKAGRSRWLGIRPTVRGTAMNPCDHPHGGGGGKS-IGNKPSQS 285
Query 64 PWGKPAL-GVKTRGKKTTDKFIVRRR 88
PWG A G KTR K +K +VR R
Sbjct 286 PWGVLAKGGYKTRRGKNVNKLLVRDR 311
> Hs21265070
Length=305
Score = 40.8 bits (94), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 18/32 (56%), Positives = 25/32 (78%), Gaps = 0/32 (0%)
Query 3 CMATIGEVGNSEHSLVQIGKAGRNRHMGRRPH 34
C+AT+G V N +H+ IGKAGRNR +G+RP+
Sbjct 240 CVATVGRVSNVDHNKRVIGKAGRNRWLGKRPN 271
> At4g36130
Length=258
Score = 40.8 bits (94), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 24/53 (45%), Positives = 31/53 (58%), Gaps = 5/53 (9%)
Query 3 CMATIGEV---GNSEHSLVQIGKAGRNRHMGRR--PHVRGSVMNPVDHPHGGG 50
C A IG+V G +E +++ G A + R P VRG MNPV+HPHGGG
Sbjct 162 CRAMIGQVAGGGRTEKPMLKAGNAYHKYRVKRNCWPKVRGVAMNPVEHPHGGG 214
> At2g18020
Length=258
Score = 40.8 bits (94), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 24/53 (45%), Positives = 31/53 (58%), Gaps = 5/53 (9%)
Query 3 CMATIGEV---GNSEHSLVQIGKAGRNRHMGRR--PHVRGSVMNPVDHPHGGG 50
C A IG+V G +E +++ G A + R P VRG MNPV+HPHGGG
Sbjct 162 CRAMIGQVAGGGRTEKPMLKAGNAYHKYRVKRNSWPKVRGVAMNPVEHPHGGG 214
> HsM7705714
Length=353
Score = 40.8 bits (94), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 18/32 (56%), Positives = 25/32 (78%), Gaps = 0/32 (0%)
Query 3 CMATIGEVGNSEHSLVQIGKAGRNRHMGRRPH 34
C+AT+G V N +H+ IGKAGRNR +G+RP+
Sbjct 240 CVATVGRVSNVDHNKRVIGKAGRNRWLGKRPN 271
> At3g51190
Length=260
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/54 (44%), Positives = 33/54 (61%), Gaps = 7/54 (12%)
Query 3 CMATIGEV---GNSEHSLVQIGKAGRNRHMGRR---PHVRGSVMNPVDHPHGGG 50
C A IG+V G +E ++ G A +++ +R P VRG MNPV+HPHGGG
Sbjct 163 CRAMIGQVAGGGRTEKPFLKAGNA-YHKYKAKRNCWPVVRGVAMNPVEHPHGGG 215
> 7294756
Length=294
Score = 37.4 bits (85), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 16/31 (51%), Positives = 20/31 (64%), Gaps = 0/31 (0%)
Query 3 CMATIGEVGNSEHSLVQIGKAGRNRHMGRRP 33
CMAT+G + N EH+ +G A R R MG RP
Sbjct 222 CMATVGRLSNPEHNKEHVGSAQRMREMGNRP 252
> CE18478
Length=260
Score = 36.6 bits (83), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 23/51 (45%), Positives = 28/51 (54%), Gaps = 5/51 (9%)
Query 5 ATIGEVGNSEHSLVQIGKAGRNRHMGRR-----PHVRGSVMNPVDHPHGGG 50
A IG V + + KAGR+ H + P VRG MNPV+HPHGGG
Sbjct 164 AMIGLVAGGGRTDKPLLKAGRSYHKYKAKRNSWPRVRGVAMNPVEHPHGGG 214
> YIL018w
Length=254
Score = 36.6 bits (83), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 20/34 (58%), Positives = 20/34 (58%), Gaps = 5/34 (14%)
Query 22 KAGRNRHMGRR-----PHVRGSVMNPVDHPHGGG 50
KAGR H R P RG MNPVDHPHGGG
Sbjct 181 KAGRAFHKYRLKRNSWPKTRGVAMNPVDHPHGGG 214
> YFR031c-a
Length=254
Score = 36.6 bits (83), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 20/34 (58%), Positives = 20/34 (58%), Gaps = 5/34 (14%)
Query 22 KAGRNRHMGRR-----PHVRGSVMNPVDHPHGGG 50
KAGR H R P RG MNPVDHPHGGG
Sbjct 181 KAGRAFHKYRLKRNSWPKTRGVAMNPVDHPHGGG 214
> SPBC839.04
Length=253
Score = 36.2 bits (82), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 20/34 (58%), Positives = 20/34 (58%), Gaps = 5/34 (14%)
Query 22 KAGRNRHMGRR-----PHVRGSVMNPVDHPHGGG 50
KAGR H R P RG MNPVDHPHGGG
Sbjct 180 KAGRAFHKYRVKRNCWPRTRGVAMNPVDHPHGGG 213
> SPBC2F12.07c
Length=253
Score = 36.2 bits (82), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 20/34 (58%), Positives = 20/34 (58%), Gaps = 5/34 (14%)
Query 22 KAGRNRHMGRR-----PHVRGSVMNPVDHPHGGG 50
KAGR H R P RG MNPVDHPHGGG
Sbjct 180 KAGRAFHKYRVKRNCWPRTRGVAMNPVDHPHGGG 213
> 7292250
Length=256
Score = 35.4 bits (80), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 20/36 (55%), Positives = 22/36 (61%), Gaps = 5/36 (13%)
Query 20 IGKAGRNRHMGRR-----PHVRGSVMNPVDHPHGGG 50
I KAGR H + P VRG MNPV+HPHGGG
Sbjct 179 ILKAGRAYHKYKVKRNSWPKVRGVAMNPVEHPHGGG 214
> Hs4506663
Length=257
Score = 33.5 bits (75), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 19/36 (52%), Positives = 21/36 (58%), Gaps = 5/36 (13%)
Query 20 IGKAGRNRHMGRR-----PHVRGSVMNPVDHPHGGG 50
I KAGR H + P VRG MNPV+HP GGG
Sbjct 179 ILKAGRAYHKYKAKRNCWPRVRGVAMNPVEHPFGGG 214
> ECU01g0310
Length=239
Score = 31.2 bits (69), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 13/18 (72%), Positives = 14/18 (77%), Gaps = 0/18 (0%)
Query 33 PHVRGSVMNPVDHPHGGG 50
P VRG MNPV+H HGGG
Sbjct 183 PRVRGVAMNPVEHIHGGG 200
> Hs17444499
Length=239
Score = 30.8 bits (68), Expect = 0.72, Method: Compositional matrix adjust.
Identities = 18/36 (50%), Positives = 20/36 (55%), Gaps = 5/36 (13%)
Query 20 IGKAGRNRHMGRR-----PHVRGSVMNPVDHPHGGG 50
I KAGR H + P VRG MN V+HP GGG
Sbjct 161 ILKAGRAHHKYKAKRNCWPRVRGVAMNLVEHPFGGG 196
> Hs21389515
Length=691
Score = 30.4 bits (67), Expect = 0.87, Method: Composition-based stats.
Identities = 11/39 (28%), Positives = 20/39 (51%), Gaps = 0/39 (0%)
Query 18 VQIGKAGRNRHMGRRPHVRGSVMNPVDHPHGGGEGKNPV 56
+Q G N ++ RRP + +P ++PH + +PV
Sbjct 650 IQEGSRDNNEYIARRPDNQEGAFDPKEYPHSAKDEAHPV 688
> At4g14250_2
Length=67
Score = 29.3 bits (64), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 19/48 (39%), Positives = 28/48 (58%), Gaps = 7/48 (14%)
Query 3 CMATIGEVGNSEHSLVQIGKAGRNRHMGRRPHVRG-SVMNPVDHPHGG 49
C IG++ +S + K R+M + VRG ++MNPV+HPHGG
Sbjct 26 CRVMIGQIASSGLT----KKLMIKRNMWAK--VRGVAMMNPVEHPHGG 67
> Hs20539203
Length=757
Score = 28.9 bits (63), Expect = 2.5, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 19/37 (51%), Gaps = 0/37 (0%)
Query 31 RRPHVRGSVMNPVDHPHGGGEGKNPVGRKAPLTPWGK 67
R P VRGS+ P + PHG + P G + T G+
Sbjct 213 RNPDVRGSLQTPQNRPHGHQSPELPEGYEQRTTVQGQ 249
> Hs4503201
Length=515
Score = 28.1 bits (61), Expect = 4.6, Method: Composition-based stats.
Identities = 14/24 (58%), Positives = 15/24 (62%), Gaps = 1/24 (4%)
Query 55 PVGRKAPLTPWGKPALG-VKTRGK 77
P G K+P PWG P LG V T GK
Sbjct 36 PKGLKSPPEPWGWPLLGHVLTLGK 59
> SPAC139.05
Length=493
Score = 27.7 bits (60), Expect = 5.4, Method: Composition-based stats.
Identities = 11/24 (45%), Positives = 14/24 (58%), Gaps = 0/24 (0%)
Query 53 KNPVGRKAPLTPWGKPALGVKTRG 76
K PVG A +TPW PA + +G
Sbjct 152 KQPVGVSALITPWNFPAAMIARKG 175
> Hs20552471
Length=398
Score = 27.3 bits (59), Expect = 6.1, Method: Composition-based stats.
Identities = 14/24 (58%), Positives = 15/24 (62%), Gaps = 1/24 (4%)
Query 55 PVGRKAPLTPWGKPALG-VKTRGK 77
P G K+P PWG P LG V T GK
Sbjct 36 PKGLKSPPEPWGWPLLGHVLTLGK 59
Lambda K H
0.315 0.137 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174483934
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40