bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_7278_orf1
Length=64
Score E
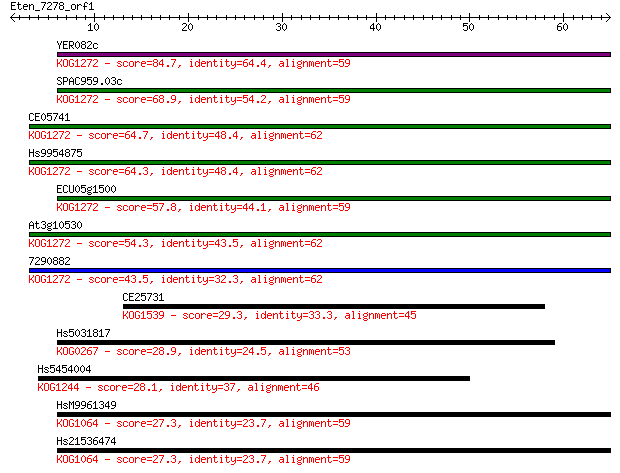
Sequences producing significant alignments: (Bits) Value
YER082c 84.7 3e-17
SPAC959.03c 68.9 2e-12
CE05741 64.7 4e-11
Hs9954875 64.3 5e-11
ECU05g1500 57.8 5e-09
At3g10530 54.3 5e-08
7290882 43.5 8e-05
CE25731 29.3 1.8
Hs5031817 28.9 2.6
Hs5454004 28.1 3.6
HsM9961349 27.3 7.5
Hs21536474 27.3 7.6
> YER082c
Length=554
Score = 84.7 bits (208), Expect = 3e-17, Method: Composition-based stats.
Identities = 38/59 (64%), Positives = 46/59 (77%), Gaps = 0/59 (0%)
Query 6 FLPYHYLMVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPNPTNAVMHLGHTKGTV 64
FLPYHYL+V+ GE G L Y+D+STGQ+ ++ RTK GP MA NP NAVMHLGH+ GTV
Sbjct 198 FLPYHYLLVTAGETGWLKYHDVSTGQLVSELRTKAGPTMAMAQNPWNAVMHLGHSNGTV 256
> SPAC959.03c
Length=520
Score = 68.9 bits (167), Expect = 2e-12, Method: Composition-based stats.
Identities = 32/59 (54%), Positives = 38/59 (64%), Gaps = 0/59 (0%)
Query 6 FLPYHYLMVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPNPTNAVMHLGHTKGTV 64
FLPYH L+ S G G L Y D+STGQ+ A+HRT G V+ NP NAV H+GH G V
Sbjct 189 FLPYHLLLTSIGNAGYLKYQDVSTGQLVAEHRTGMGASHVLHQNPHNAVEHVGHANGQV 247
> CE05741
Length=580
Score = 64.7 bits (156), Expect = 4e-11, Method: Composition-based stats.
Identities = 30/62 (48%), Positives = 38/62 (61%), Gaps = 0/62 (0%)
Query 3 RRPFLPYHYLMVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPNPTNAVMHLGHTKG 62
R FLP+H+L+V + LNY D+S G+ A TK G VM NP NA++H GHT G
Sbjct 253 RLEFLPHHFLLVGSSRNSFLNYVDVSVGKQVASFATKSGTLDVMCQNPANAIIHTGHTNG 312
Query 63 TV 64
TV
Sbjct 313 TV 314
> Hs9954875
Length=610
Score = 64.3 bits (155), Expect = 5e-11, Method: Composition-based stats.
Identities = 30/62 (48%), Positives = 40/62 (64%), Gaps = 0/62 (0%)
Query 3 RRPFLPYHYLMVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPNPTNAVMHLGHTKG 62
R FLP+H+L+ + E G L Y D+S G+I A + G VM+ NP NAV+HLGH+ G
Sbjct 280 RLEFLPFHFLLATASETGFLTYLDVSVGKIVAALNARAGRLDVMSQNPYNAVIHLGHSNG 339
Query 63 TV 64
TV
Sbjct 340 TV 341
> ECU05g1500
Length=435
Score = 57.8 bits (138), Expect = 5e-09, Method: Composition-based stats.
Identities = 26/59 (44%), Positives = 37/59 (62%), Gaps = 0/59 (0%)
Query 6 FLPYHYLMVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPNPTNAVMHLGHTKGTV 64
+LPYHYL+V+ + G L Y DISTG+I ++ + M NP NA++H G+ KG V
Sbjct 160 YLPYHYLLVAASDKGFLRYQDISTGKIVSQIYLRNREVTSMKQNPMNAIIHTGNVKGVV 218
> At3g10530
Length=548
Score = 54.3 bits (129), Expect = 5e-08, Method: Composition-based stats.
Identities = 27/62 (43%), Positives = 38/62 (61%), Gaps = 0/62 (0%)
Query 3 RRPFLPYHYLMVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPNPTNAVMHLGHTKG 62
R FL H+L+ S G+L+Y D++ G + A RT +G VM NP N+V+ LGH+ G
Sbjct 214 RLRFLKNHFLLASVNMSGQLHYQDVTHGGMVASIRTGKGRTDVMEVNPYNSVVGLGHSGG 273
Query 63 TV 64
TV
Sbjct 274 TV 275
> 7290882
Length=609
Score = 43.5 bits (101), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 33/62 (53%), Gaps = 0/62 (0%)
Query 3 RRPFLPYHYLMVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPNPTNAVMHLGHTKG 62
R FLPYH+L+ + G ++ D+S G++ T G ++ NP N V+ +G +G
Sbjct 258 RLDFLPYHFLLAAGNSAGYASWLDVSIGELVGNFNTGLGDIRMLRHNPRNGVLCIGGGRG 317
Query 63 TV 64
V
Sbjct 318 VV 319
> CE25731
Length=897
Score = 29.3 bits (64), Expect = 1.8, Method: Composition-based stats.
Identities = 15/45 (33%), Positives = 26/45 (57%), Gaps = 0/45 (0%)
Query 13 MVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPNPTNAVMHL 57
++ST G + ++ +ST I AK K G+ AP P+N+++ L
Sbjct 487 LMSTDAQGHILFWSLSTKAINAKMFKKDVRLGICAPCPSNSLVAL 531
> Hs5031817
Length=655
Score = 28.9 bits (63), Expect = 2.6, Method: Composition-based stats.
Identities = 13/53 (24%), Positives = 25/53 (47%), Gaps = 0/53 (0%)
Query 6 FLPYHYLMVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPNPTNAVMHLG 58
F P YL+ S G + ++D+ Q+ ++ + GP + NP ++ G
Sbjct 197 FHPNEYLLASGSSDGTIRFWDLEKFQVVSRIEGEPGPVRSVLFNPDGCCLYSG 249
> Hs5454004
Length=391
Score = 28.1 bits (61), Expect = 3.6, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 24/50 (48%), Gaps = 4/50 (8%)
Query 4 RPFLPYHY----LMVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPN 49
RP L YHY L GE E + Q + + ++K+GP G+ PN
Sbjct 221 RPGLSYHYAHSHLAEEEGEDKEDSQPPTPVSQRSEEQKSKKGPDGLALPN 270
> HsM9961349
Length=3027
Score = 27.3 bits (59), Expect = 7.5, Method: Composition-based stats.
Identities = 14/59 (23%), Positives = 25/59 (42%), Gaps = 0/59 (0%)
Query 6 FLPYHYLMVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPNPTNAVMHLGHTKGTV 64
+ P H L++S G G +D+ Q ++ P +A +PT G +G +
Sbjct 2894 YAPKHQLLISGGRKGFTYVFDLCQRQQRQLFQSHDSPVKAVAVDPTEEYFVTGSAEGNI 2952
> Hs21536474
Length=3027
Score = 27.3 bits (59), Expect = 7.6, Method: Composition-based stats.
Identities = 14/59 (23%), Positives = 25/59 (42%), Gaps = 0/59 (0%)
Query 6 FLPYHYLMVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPNPTNAVMHLGHTKGTV 64
+ P H L++S G G +D+ Q ++ P +A +PT G +G +
Sbjct 2894 YAPKHQLLISGGRKGFTYVFDLCQRQQRQLFQSHDSPVKAVAVDPTEEYFVTGSAEGNI 2952
Lambda K H
0.321 0.138 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1169706042
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40