bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_7494_orf1
Length=90
Score E
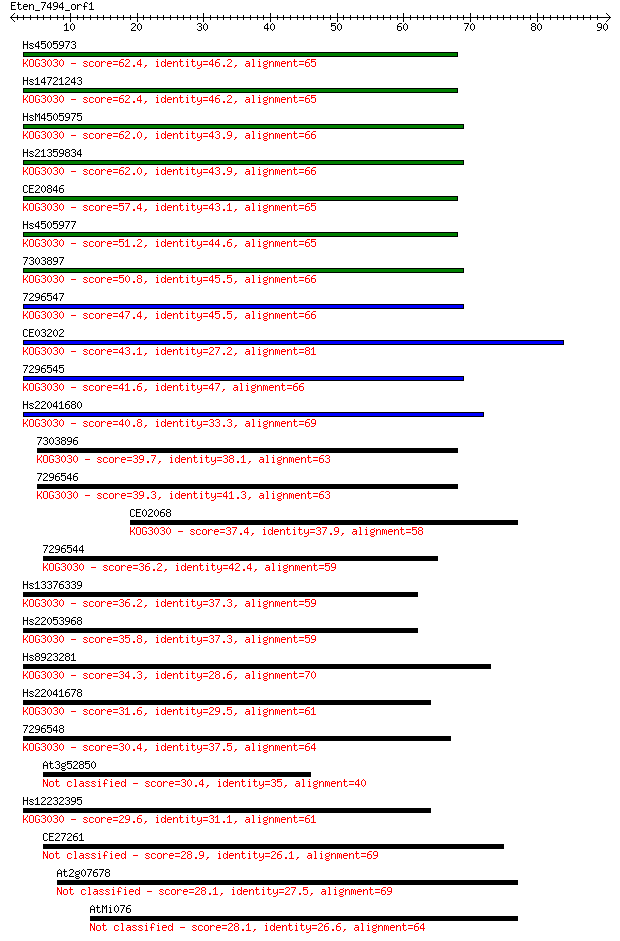
Sequences producing significant alignments: (Bits) Value
Hs4505973 62.4 2e-10
Hs14721243 62.4 2e-10
HsM4505975 62.0 2e-10
Hs21359834 62.0 2e-10
CE20846 57.4 6e-09
Hs4505977 51.2 4e-07
7303897 50.8 7e-07
7296547 47.4 6e-06
CE03202 43.1 1e-04
7296545 41.6 3e-04
Hs22041680 40.8 6e-04
7303896 39.7 0.001
7296546 39.3 0.002
CE02068 37.4 0.006
7296544 36.2 0.016
Hs13376339 36.2 0.017
Hs22053968 35.8 0.018
Hs8923281 34.3 0.054
Hs22041678 31.6 0.37
7296548 30.4 0.91
At3g52850 30.4 0.93
Hs12232395 29.6 1.2
CE27261 28.9 2.3
At2g07678 28.1 3.8
AtMi076 28.1 3.9
> Hs4505973
Length=289
Score = 62.4 bits (150), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 30/65 (46%), Positives = 40/65 (61%), Gaps = 5/65 (7%)
Query 3 LAVCQPNWGSLKCSDSTGDLYVDQYECRGADQAAIKEARLSFFSAHSSNSSCAMLYTIIY 62
L VC P+W + CSD Y++ Y CRG + +KE RLSF+S HSS S ML+ +Y
Sbjct 136 LDVCDPDWSKINCSDG----YIEYYICRG-NAERVKEGRLSFYSGHSSFSMYCMLFVALY 190
Query 63 LQCRL 67
LQ R+
Sbjct 191 LQARM 195
> Hs14721243
Length=285
Score = 62.4 bits (150), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 30/65 (46%), Positives = 40/65 (61%), Gaps = 5/65 (7%)
Query 3 LAVCQPNWGSLKCSDSTGDLYVDQYECRGADQAAIKEARLSFFSAHSSNSSCAMLYTIIY 62
L VC P+W + CSD Y++ Y CRG + +KE RLSF+S HSS S ML+ +Y
Sbjct 132 LDVCDPDWSKINCSDG----YIEYYICRG-NAERVKEGRLSFYSGHSSFSMYCMLFVALY 186
Query 63 LQCRL 67
LQ R+
Sbjct 187 LQARM 191
> HsM4505975
Length=311
Score = 62.0 bits (149), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 29/66 (43%), Positives = 42/66 (63%), Gaps = 5/66 (7%)
Query 3 LAVCQPNWGSLKCSDSTGDLYVDQYECRGADQAAIKEARLSFFSAHSSNSSCAMLYTIIY 62
L+VC P++ + CS+ Y+ Y CRG D + ++EAR SFFS H+S S MLY ++Y
Sbjct 159 LSVCNPDFSQINCSEG----YIQNYRCRG-DDSKVQEARKSFFSGHASFSMYTMLYLVLY 213
Query 63 LQCRLS 68
LQ R +
Sbjct 214 LQARFT 219
> Hs21359834
Length=311
Score = 62.0 bits (149), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 29/66 (43%), Positives = 42/66 (63%), Gaps = 5/66 (7%)
Query 3 LAVCQPNWGSLKCSDSTGDLYVDQYECRGADQAAIKEARLSFFSAHSSNSSCAMLYTIIY 62
L+VC P++ + CS+ Y+ Y CRG D + ++EAR SFFS H+S S MLY ++Y
Sbjct 159 LSVCNPDFSQINCSEG----YIQNYRCRG-DDSKVQEARKSFFSGHASFSMYTMLYLVLY 213
Query 63 LQCRLS 68
LQ R +
Sbjct 214 LQARFT 219
> CE20846
Length=346
Score = 57.4 bits (137), Expect = 6e-09, Method: Composition-based stats.
Identities = 28/65 (43%), Positives = 39/65 (60%), Gaps = 2/65 (3%)
Query 3 LAVCQPNWGSLKCSDSTGDLYVDQYECRGADQAAIKEARLSFFSAHSSNSSCAMLYTIIY 62
+ VC+P+ G CS DLY+ ++C D I EA+LSF+S HS+ S A +T +Y
Sbjct 142 MDVCRPDIGYQTCSQP--DLYITDFKCTTTDTKKIHEAQLSFYSGHSAFSFYAAWFTSLY 199
Query 63 LQCRL 67
LQ RL
Sbjct 200 LQARL 204
> Hs4505977
Length=288
Score = 51.2 bits (121), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 29/66 (43%), Positives = 40/66 (60%), Gaps = 7/66 (10%)
Query 3 LAVCQPNWGSLKCSDSTGDLYVD-QYECRGADQAAIKEARLSFFSAHSSNSSCAMLYTII 61
LAVC P+W + CS +YV + CRG + A + EARLSF+S HSS M++ +
Sbjct 128 LAVCDPDWSRVNCS-----VYVQLEKVCRG-NPADVTEARLSFYSGHSSFGMYCMVFLAL 181
Query 62 YLQCRL 67
Y+Q RL
Sbjct 182 YVQARL 187
> 7303897
Length=372
Score = 50.8 bits (120), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 30/71 (42%), Positives = 43/71 (60%), Gaps = 6/71 (8%)
Query 3 LAVCQPNW--GSLKCSDS-TGDLYVDQYECRGADQAA--IKEARLSFFSAHSSNSSCAML 57
+AVCQP GS C D+ Y+ ++ C+G +A +KE RLSF S HSS + AM+
Sbjct 215 IAVCQPQMADGS-TCDDAINAGKYIQEFTCKGVGSSARMLKEMRLSFPSGHSSFTFFAMV 273
Query 58 YTIIYLQCRLS 68
Y +YLQ R++
Sbjct 274 YLALYLQARMT 284
> 7296547
Length=340
Score = 47.4 bits (111), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 30/71 (42%), Positives = 43/71 (60%), Gaps = 6/71 (8%)
Query 3 LAVCQPNW--GSLKCSDSTG-DLYVDQYECRGADQAA--IKEARLSFFSAHSSNSSCAML 57
LAVCQP GS+ CSD Y++ Y+C G +++ARLSF S HSS + AM+
Sbjct 164 LAVCQPQIADGSM-CSDPVNLHRYMENYDCAGEGFTVEDVRQARLSFPSGHSSLAFYAMI 222
Query 58 YTIIYLQCRLS 68
Y +YLQ +++
Sbjct 223 YVALYLQRKIT 233
> CE03202
Length=318
Score = 43.1 bits (100), Expect = 1e-04, Method: Composition-based stats.
Identities = 22/81 (27%), Positives = 40/81 (49%), Gaps = 1/81 (1%)
Query 3 LAVCQPNWGSLKCSDSTGDLYVDQYECRGADQAAIKEARLSFFSAHSSNSSCAMLYTIIY 62
+VC+P+W + C+D + C + I+ AR SF S H++ + L+ IY
Sbjct 172 FSVCKPDWSKVDCTDKQSFIDSSDLVCTNPNPRKIRTARTSFPSGHTAAAFHVFLFVYIY 231
Query 63 LQCRLSKASAGPLMATFRQRL 83
L+ R+++ + + T R L
Sbjct 232 LR-RMAENTGIKEIITIRNIL 251
> 7296545
Length=341
Score = 41.6 bits (96), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 31/71 (43%), Positives = 37/71 (52%), Gaps = 6/71 (8%)
Query 3 LAVCQPNW--GSLKCSDSTG-DLYVDQYECRGADQA--AIKEARLSFFSAHSSNSSCAML 57
LAVCQP GS C D+ Y+D + C A+ KE SF S H+S + AML
Sbjct 137 LAVCQPMLPDGS-SCQDAQNLGRYIDSFTCSNANMTDYQFKELYQSFPSGHASMAMYAML 195
Query 58 YTIIYLQCRLS 68
Y IYLQ LS
Sbjct 196 YLAIYLQAALS 206
> Hs22041680
Length=763
Score = 40.8 bits (94), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 39/71 (54%), Gaps = 5/71 (7%)
Query 3 LAVCQPNWGSLK--CSDSTGDLYVDQYECRGADQAAIKEARLSFFSAHSSNSSCAMLYTI 60
L VC+PN+ SL C +++ Y+ + C G+D I R SF S H++ ++ A +Y
Sbjct 208 LTVCKPNYTSLNVSCKENS---YIVEDICSGSDLTVINSGRKSFPSQHATLAAFAAVYVS 264
Query 61 IYLQCRLSKAS 71
+Y L+ +S
Sbjct 265 MYFNSTLTDSS 275
> 7303896
Length=246
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 37/67 (55%), Gaps = 4/67 (5%)
Query 5 VCQPNWG-SLKCSDSTGDL-YVDQYECRGADQAA--IKEARLSFFSAHSSNSSCAMLYTI 60
+CQP CSD Y++++ C D + +K+ RLSF S H+S + +MLY +
Sbjct 123 LCQPVMKDGTTCSDPINAARYIEEFTCAAVDITSKQLKDMRLSFPSGHASFACYSMLYLV 182
Query 61 IYLQCRL 67
IYL R+
Sbjct 183 IYLHRRM 189
> 7296546
Length=305
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 26/71 (36%), Positives = 34/71 (47%), Gaps = 8/71 (11%)
Query 5 VCQPNWGS---LKCSDSTGD---LYVDQYECR--GADQAAIKEARLSFFSAHSSNSSCAM 56
+CQP WG+ CSD T LY++ + C A Q + R SF S S + AM
Sbjct 145 ICQPTWGTEGGESCSDVTAQNSTLYLEDFSCTEFAASQDLLALVRHSFPSGFVSTTCYAM 204
Query 57 LYTIIYLQCRL 67
+ I Y Q RL
Sbjct 205 GFLIFYSQARL 215
> CE02068
Length=341
Score = 37.4 bits (85), Expect = 0.006, Method: Composition-based stats.
Identities = 22/60 (36%), Positives = 33/60 (55%), Gaps = 8/60 (13%)
Query 19 TGD--LYVDQYECRGADQAAIKEARLSFFSAHSSNSSCAMLYTIIYLQCRLSKASAGPLM 76
TGD Y+ Y C G + + EAR SF+S HS+ S ++ +Y+Q RL GP++
Sbjct 167 TGDSHRYITDYTCTGPPELVL-EARKSFYSGHSAVSLYCATWSALYIQARL-----GPVL 220
> 7296544
Length=334
Score = 36.2 bits (82), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 25/64 (39%), Positives = 36/64 (56%), Gaps = 6/64 (9%)
Query 6 CQPNW--GSLKCSD-STGDLYVDQYECRGADQAA--IKEARLSFFSAHSSNSSCAMLYTI 60
CQP GS CSD +LYV+Q+ C + + I+E +SF SAHSS S +M+
Sbjct 98 CQPRLDDGS-SCSDLQNAELYVEQFHCTNNNLSTRQIRELHVSFPSAHSSLSFYSMVLLA 156
Query 61 IYLQ 64
+Y+
Sbjct 157 LYVH 160
> Hs13376339
Length=746
Score = 36.2 bits (82), Expect = 0.017, Method: Composition-based stats.
Identities = 22/59 (37%), Positives = 32/59 (54%), Gaps = 1/59 (1%)
Query 3 LAVCQPNWGSLKCSDSTGDLYVDQYECRGADQAAIKEARLSFFSAHSSNSSCAMLYTII 61
L VC+PN+ L S + Y+ Q C G D AI AR +F S H++ S+ A +Y +
Sbjct 163 LTVCKPNYTLLGTSCEV-NPYITQDICSGHDIHAILSARKTFPSQHATLSAFAAVYVSV 220
> Hs22053968
Length=686
Score = 35.8 bits (81), Expect = 0.018, Method: Composition-based stats.
Identities = 22/59 (37%), Positives = 32/59 (54%), Gaps = 1/59 (1%)
Query 3 LAVCQPNWGSLKCSDSTGDLYVDQYECRGADQAAIKEARLSFFSAHSSNSSCAMLYTII 61
L VC+PN+ L S + Y+ Q C G D AI AR +F S H++ S+ A +Y +
Sbjct 103 LTVCKPNYTLLGTSCEV-NPYITQDICSGHDIHAILSARKTFPSQHATLSAFAAVYVSV 160
> Hs8923281
Length=325
Score = 34.3 bits (77), Expect = 0.054, Method: Composition-based stats.
Identities = 20/70 (28%), Positives = 34/70 (48%), Gaps = 2/70 (2%)
Query 3 LAVCQPNWGSLKCSDSTGDLYVDQYECRGADQAAIKEARLSFFSAHSSNSSCAMLYTIIY 62
L VC+PN+ S C +++ D I++AR SF S H++ S + LY +Y
Sbjct 157 LTVCKPNYTSADCQ--AHHQFINNGNICTGDLEVIEKARRSFPSKHAALSIYSALYATMY 214
Query 63 LQCRLSKASA 72
+ + S+
Sbjct 215 ITSTIKTKSS 224
> Hs22041678
Length=321
Score = 31.6 bits (70), Expect = 0.37, Method: Composition-based stats.
Identities = 18/61 (29%), Positives = 31/61 (50%), Gaps = 2/61 (3%)
Query 3 LAVCQPNWGSLKCSDSTGDLYVDQYECRGADQAAIKEARLSFFSAHSSNSSCAMLYTIIY 62
LA+C+PN+ +L C T ++ E + I AR +F S ++ S A +Y +Y
Sbjct 152 LALCKPNYTALGCQQYTQ--FISGEEACTGNPDLIMRARKTFPSKEAALSVYAAMYLTMY 209
Query 63 L 63
+
Sbjct 210 I 210
> 7296548
Length=305
Score = 30.4 bits (67), Expect = 0.91, Method: Compositional matrix adjust.
Identities = 24/73 (32%), Positives = 39/73 (53%), Gaps = 10/73 (13%)
Query 3 LAVCQPNW--GSLKCSDST--GDL-YVDQYECRG----ADQAAIKEARLSFFSAHSSNSS 53
AVC P++ GS C D + G L Y YECR A + I++ +SF S HS+ +
Sbjct 130 FAVCSPHFPDGS-SCLDESHRGALKYHTDYECRPNLSQATEEMIRDVNVSFPSGHSAMAF 188
Query 54 CAMLYTIIYLQCR 66
+++ ++L+ R
Sbjct 189 YGLVFVALHLRRR 201
> At3g52850
Length=623
Score = 30.4 bits (67), Expect = 0.93, Method: Composition-based stats.
Identities = 14/41 (34%), Positives = 23/41 (56%), Gaps = 1/41 (2%)
Query 6 CQPNWGSLKCSDSTGDLYVDQYE-CRGADQAAIKEARLSFF 45
C+ WGS +CS S G LY+ +++ C G+ + + SF
Sbjct 529 CKNTWGSYECSCSNGLLYMREHDTCIGSGKVGTTKLSWSFL 569
> Hs12232395
Length=343
Score = 29.6 bits (65), Expect = 1.2, Method: Composition-based stats.
Identities = 19/67 (28%), Positives = 34/67 (50%), Gaps = 7/67 (10%)
Query 3 LAVCQPNWGSLKCSDSTGD------LYVDQYECRGADQAAIKEARLSFFSAHSSNSSCAM 56
L+VC+PN+ +L C + D DQ C G+ + + AR +F ++ + A+
Sbjct 159 LSVCRPNYTALGCLPPSPDRPGPDRFVTDQGACAGS-PSLVAAARRAFPCKDAALCAYAV 217
Query 57 LYTIIYL 63
YT +Y+
Sbjct 218 TYTAMYV 224
> CE27261
Length=4155
Score = 28.9 bits (63), Expect = 2.3, Method: Composition-based stats.
Identities = 18/69 (26%), Positives = 32/69 (46%), Gaps = 13/69 (18%)
Query 6 CQPNWGSLKCSDSTGDLYVDQYECRGADQAAIKEARLSFFSAHSSNSSCAMLYTIIYLQC 65
C+ ++ KCS +VD + D+ A+ ++ L + HS S+C Y C
Sbjct 2858 CKASFNGFKCS------FVDDF-----DEQALTQSFLD--ATHSDQSNCIFFPEGTYQLC 2904
Query 66 RLSKASAGP 74
++A+ GP
Sbjct 2905 SKAEATKGP 2913
> At2g07678
Length=245
Score = 28.1 bits (61), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 19/69 (27%), Positives = 34/69 (49%), Gaps = 4/69 (5%)
Query 8 PNWGSLKCSDSTGDLYVDQYECRGADQAAIKEARLSFFSAHSSNSSCAMLYTIIYLQCRL 67
P+ G LKC D+TGD + + + + +S + S NS A+ + I++ L
Sbjct 85 PSSG-LKCLDTTGDFLIKNVLHKRYESV---QKNISNALSSSINSRTAVFFCILFSITVL 140
Query 68 SKASAGPLM 76
+ + GPL+
Sbjct 141 MEIAPGPLL 149
> AtMi076
Length=215
Score = 28.1 bits (61), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 17/64 (26%), Positives = 31/64 (48%), Gaps = 3/64 (4%)
Query 13 LKCSDSTGDLYVDQYECRGADQAAIKEARLSFFSAHSSNSSCAMLYTIIYLQCRLSKASA 72
LKC D+TGD + + + + +S + S NS A+ + I++ L + +
Sbjct 89 LKCLDTTGDFLIKNVLHKRYESV---QKNISNALSSSINSRTAVFFCILFSITVLMEIAP 145
Query 73 GPLM 76
GPL+
Sbjct 146 GPLL 149
Lambda K H
0.320 0.127 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1177758614
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40