bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_7587_orf2
Length=66
Score E
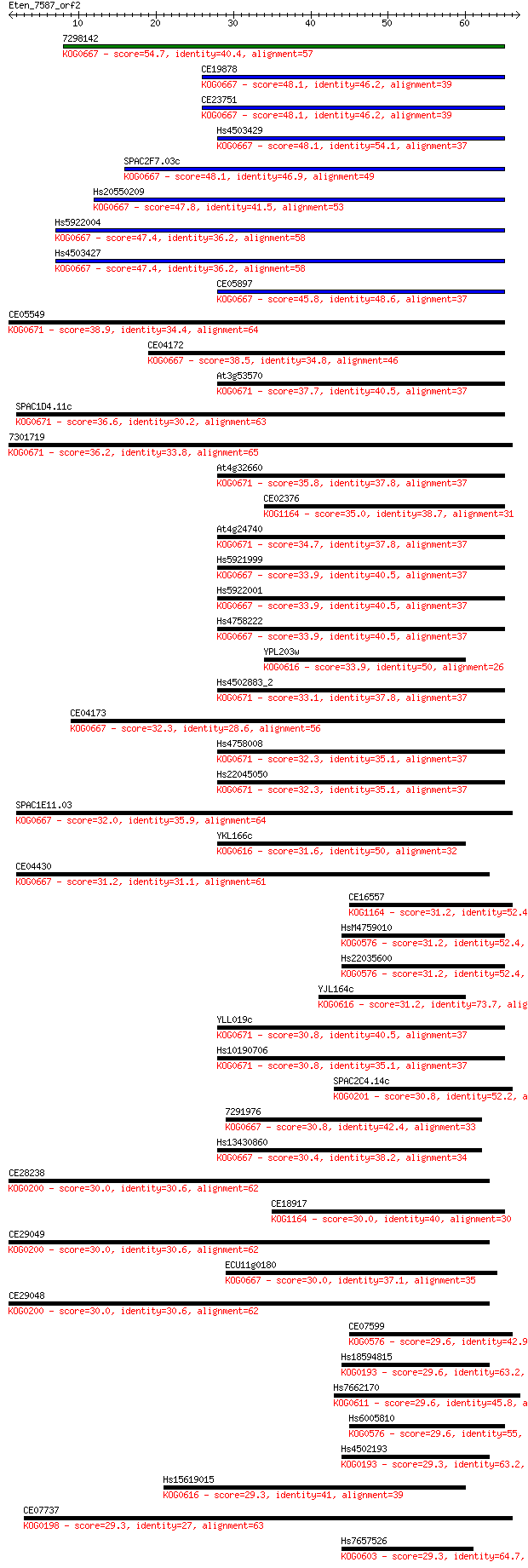
Sequences producing significant alignments: (Bits) Value
7298142 54.7 5e-08
CE19878 48.1 3e-06
CE23751 48.1 4e-06
Hs4503429 48.1 4e-06
SPAC2F7.03c 48.1 4e-06
Hs20550209 47.8 5e-06
Hs5922004 47.4 6e-06
Hs4503427 47.4 6e-06
CE05897 45.8 2e-05
CE05549 38.9 0.002
CE04172 38.5 0.003
At3g53570 37.7 0.005
SPAC1D4.11c 36.6 0.011
7301719 36.2 0.013
At4g32660 35.8 0.017
CE02376 35.0 0.034
At4g24740 34.7 0.048
Hs5921999 33.9 0.066
Hs5922001 33.9 0.068
Hs4758222 33.9 0.068
YPL203w 33.9 0.083
Hs4502883_2 33.1 0.12
CE04173 32.3 0.21
Hs4758008 32.3 0.23
Hs22045050 32.3 0.23
SPAC1E11.03 32.0 0.27
YKL166c 31.6 0.34
CE04430 31.2 0.43
CE16557 31.2 0.45
HsM4759010 31.2 0.50
Hs22035600 31.2 0.50
YJL164c 31.2 0.53
YLL019c 30.8 0.56
Hs10190706 30.8 0.60
SPAC2C4.14c 30.8 0.70
7291976 30.8 0.70
Hs13430860 30.4 0.76
CE28238 30.0 0.96
CE18917 30.0 0.96
CE29049 30.0 1.0
ECU11g0180 30.0 1.1
CE29048 30.0 1.1
CE07599 29.6 1.5
Hs18594815 29.6 1.6
Hs7662170 29.6 1.6
Hs6005810 29.6 1.6
Hs4502193 29.3 1.6
Hs15619015 29.3 1.6
CE07737 29.3 1.8
Hs7657526 29.3 1.8
> 7298142
Length=722
Score = 54.7 bits (130), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 23/57 (40%), Positives = 38/57 (66%), Gaps = 0/57 (0%)
Query 8 SSSNRQLMNATSNSACSYFCDADGDYIVSQNDHICYRFQLLQTLGTGSFGQVFRALD 64
+S N T+N+ + D +G+Y + ++DHI +R+++L+ +G GSFGQV RALD
Sbjct 161 ASKNYNKPAPTANTTNLGYDDDNGNYKIIEHDHIAFRYEILEVIGKGSFGQVIRALD 217
> CE19878
Length=817
Score = 48.1 bits (113), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 18/39 (46%), Positives = 28/39 (71%), Gaps = 0/39 (0%)
Query 26 FCDADGDYIVSQNDHICYRFQLLQTLGTGSFGQVFRALD 64
+ D +G Y + +DHI YR+++L+ +G GSFGQV +A D
Sbjct 442 YDDENGSYQLVVHDHIAYRYEVLKVIGKGSFGQVIKAFD 480
> CE23751
Length=796
Score = 48.1 bits (113), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 18/39 (46%), Positives = 28/39 (71%), Gaps = 0/39 (0%)
Query 26 FCDADGDYIVSQNDHICYRFQLLQTLGTGSFGQVFRALD 64
+ D +G Y + +DHI YR+++L+ +G GSFGQV +A D
Sbjct 442 YDDENGSYQLVVHDHIAYRYEVLKVIGKGSFGQVIKAFD 480
> Hs4503429
Length=553
Score = 48.1 bits (113), Expect = 4e-06, Method: Composition-based stats.
Identities = 20/37 (54%), Positives = 26/37 (70%), Gaps = 0/37 (0%)
Query 28 DADGDYIVSQNDHICYRFQLLQTLGTGSFGQVFRALD 64
DADG YI DH+ YR+++L+ +G GSFGQV R D
Sbjct 157 DADGAYIHVPRDHLAYRYEVLKIIGKGSFGQVARVYD 193
> SPAC2F7.03c
Length=1087
Score = 48.1 bits (113), Expect = 4e-06, Method: Composition-based stats.
Identities = 23/50 (46%), Positives = 33/50 (66%), Gaps = 1/50 (2%)
Query 16 NATSNSACSY-FCDADGDYIVSQNDHICYRFQLLQTLGTGSFGQVFRALD 64
+A N++ ++ F D GDY V DHI YR++++ LG GSFGQV R +D
Sbjct 669 SADENTSSNFGFDDERGDYKVVLGDHIAYRYEVVDFLGKGSFGQVLRCID 718
> Hs20550209
Length=520
Score = 47.8 bits (112), Expect = 5e-06, Method: Composition-based stats.
Identities = 22/53 (41%), Positives = 34/53 (64%), Gaps = 0/53 (0%)
Query 12 RQLMNATSNSACSYFCDADGDYIVSQNDHICYRFQLLQTLGTGSFGQVFRALD 64
++L A + + F D G Y+ +DHI YR+++L+T+G GSFGQV + LD
Sbjct 71 KKLDTAPEKFSKTSFDDEHGFYLKVLHDHIAYRYEVLETIGKGSFGQVAKCLD 123
> Hs5922004
Length=601
Score = 47.4 bits (111), Expect = 6e-06, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 35/58 (60%), Gaps = 2/58 (3%)
Query 7 MSSSNRQLMNATSNSACSYFCDADGDYIVSQNDHICYRFQLLQTLGTGSFGQVFRALD 64
+++ RQ M N+ + D G Y+ +DH+ YR+++L+ +G GSFGQV +A D
Sbjct 186 LNAKKRQGMTGGPNNGG--YDDDQGSYVQVPHDHVAYRYEVLKVIGKGSFGQVVKAYD 241
> Hs4503427
Length=528
Score = 47.4 bits (111), Expect = 6e-06, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 35/58 (60%), Gaps = 2/58 (3%)
Query 7 MSSSNRQLMNATSNSACSYFCDADGDYIVSQNDHICYRFQLLQTLGTGSFGQVFRALD 64
+++ RQ M N+ + D G Y+ +DH+ YR+++L+ +G GSFGQV +A D
Sbjct 113 LNAKKRQGMTGGPNNGG--YDDDQGSYVQVPHDHVAYRYEVLKVIGKGSFGQVVKAYD 168
> CE05897
Length=508
Score = 45.8 bits (107), Expect = 2e-05, Method: Composition-based stats.
Identities = 18/37 (48%), Positives = 27/37 (72%), Gaps = 0/37 (0%)
Query 28 DADGDYIVSQNDHICYRFQLLQTLGTGSFGQVFRALD 64
D +G Y + +DHI YR+++L+ +G GSFGQV +A D
Sbjct 150 DENGSYQLVVHDHIAYRYEVLKVIGKGSFGQVIKAFD 186
> CE05549
Length=903
Score = 38.9 bits (89), Expect = 0.002, Method: Composition-based stats.
Identities = 22/66 (33%), Positives = 33/66 (50%), Gaps = 2/66 (3%)
Query 1 RPRRQRMSSSNR--QLMNATSNSACSYFCDADGDYIVSQNDHICYRFQLLQTLGTGSFGQ 58
+P R + +S ++ S+ + D DG I S+ D I RF + TLG G+FG+
Sbjct 505 KPSRSGLQASQARPKVPEIVSSQRTQHQDDKDGHLIYSKGDFILNRFTIYDTLGEGTFGK 564
Query 59 VFRALD 64
V R D
Sbjct 565 VVRVND 570
> CE04172
Length=450
Score = 38.5 bits (88), Expect = 0.003, Method: Composition-based stats.
Identities = 16/46 (34%), Positives = 24/46 (52%), Gaps = 0/46 (0%)
Query 19 SNSACSYFCDADGDYIVSQNDHICYRFQLLQTLGTGSFGQVFRALD 64
+N F +G Y + DHI YR+ + + L +G + QVFR D
Sbjct 105 TNHELGIFTLPNGSYCFKEGDHISYRYSIDEVLSSGYYAQVFRCTD 150
> At3g53570
Length=467
Score = 37.7 bits (86), Expect = 0.005, Method: Composition-based stats.
Identities = 15/37 (40%), Positives = 21/37 (56%), Gaps = 0/37 (0%)
Query 28 DADGDYIVSQNDHICYRFQLLQTLGTGSFGQVFRALD 64
D DG Y+ D + R+Q+L +G G+FGQV D
Sbjct 98 DKDGHYVFVVGDTLTPRYQILSKMGEGTFGQVLECFD 134
> SPAC1D4.11c
Length=690
Score = 36.6 bits (83), Expect = 0.011, Method: Composition-based stats.
Identities = 19/63 (30%), Positives = 33/63 (52%), Gaps = 0/63 (0%)
Query 2 PRRQRMSSSNRQLMNATSNSACSYFCDADGDYIVSQNDHICYRFQLLQTLGTGSFGQVFR 61
P +++ N +++ T+ S F D DG Y V N R+ +++ LG G+FG+V +
Sbjct 319 PPYKKVDRVNVPVVHDTTAFDPSTFDDDDGHYKVVPNSKFANRYTVVRLLGHGTFGKVIQ 378
Query 62 ALD 64
D
Sbjct 379 CYD 381
> 7301719
Length=517
Score = 36.2 bits (82), Expect = 0.013, Method: Composition-based stats.
Identities = 22/66 (33%), Positives = 36/66 (54%), Gaps = 3/66 (4%)
Query 1 RPRRQRMSS-SNRQLMNATSNSACSYFCDADGDYIVSQNDHICYRFQLLQTLGTGSFGQV 59
R + +R S +NRQ T+ DADG I D + +R++++ TLG G+FG+V
Sbjct 127 RAKDERDSGRNNRQSQAKTAKPVIQD--DADGHLIYHTGDILHHRYKIMATLGEGTFGRV 184
Query 60 FRALDL 65
+ D+
Sbjct 185 VKVKDM 190
> At4g32660
Length=400
Score = 35.8 bits (81), Expect = 0.017, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 23/37 (62%), Gaps = 0/37 (0%)
Query 28 DADGDYIVSQNDHICYRFQLLQTLGTGSFGQVFRALD 64
D DG Y+ S D++ R+++L +G G+FG+V D
Sbjct 54 DRDGHYVFSLRDNLTPRYKILSKMGEGTFGRVLECWD 90
> CE02376
Length=321
Score = 35.0 bits (79), Expect = 0.034, Method: Composition-based stats.
Identities = 12/31 (38%), Positives = 25/31 (80%), Gaps = 0/31 (0%)
Query 34 IVSQNDHICYRFQLLQTLGTGSFGQVFRALD 64
++++N+ I R++L+ +G+GSFG+V++A D
Sbjct 25 LLAKNNRIAKRYKLIHKVGSGSFGEVWQATD 55
> At4g24740
Length=427
Score = 34.7 bits (78), Expect = 0.048, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 28 DADGDYIVSQNDHICYRFQLLQTLGTGSFGQVFRALD 64
D DG YI D + R+++ +G G+FGQV D
Sbjct 81 DKDGHYIFELGDDLTPRYKIYSKMGEGTFGQVLECWD 117
> Hs5921999
Length=589
Score = 33.9 bits (76), Expect = 0.066, Method: Composition-based stats.
Identities = 15/37 (40%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 28 DADGDYIVSQNDHICYRFQLLQTLGTGSFGQVFRALD 64
D + DYIV + R+++ +G GSFGQV +A D
Sbjct 94 DDNHDYIVRSGERWLERYEIDSLIGKGSFGQVVKAYD 130
> Hs5922001
Length=601
Score = 33.9 bits (76), Expect = 0.068, Method: Composition-based stats.
Identities = 15/37 (40%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 28 DADGDYIVSQNDHICYRFQLLQTLGTGSFGQVFRALD 64
D + DYIV + R+++ +G GSFGQV +A D
Sbjct 94 DDNHDYIVRSGERWLERYEIDSLIGKGSFGQVVKAYD 130
> Hs4758222
Length=629
Score = 33.9 bits (76), Expect = 0.068, Method: Composition-based stats.
Identities = 15/37 (40%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 28 DADGDYIVSQNDHICYRFQLLQTLGTGSFGQVFRALD 64
D + DYIV + R+++ +G GSFGQV +A D
Sbjct 94 DDNHDYIVRSGERWLERYEIDSLIGKGSFGQVVKAYD 130
> YPL203w
Length=380
Score = 33.9 bits (76), Expect = 0.083, Method: Composition-based stats.
Identities = 13/26 (50%), Positives = 21/26 (80%), Gaps = 0/26 (0%)
Query 34 IVSQNDHICYRFQLLQTLGTGSFGQV 59
+VS+ + + FQ+++TLGTGSFG+V
Sbjct 59 LVSKGKYTLHDFQIMRTLGTGSFGRV 84
> Hs4502883_2
Length=363
Score = 33.1 bits (74), Expect = 0.12, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 24/37 (64%), Gaps = 0/37 (0%)
Query 28 DADGDYIVSQNDHICYRFQLLQTLGTGSFGQVFRALD 64
DA+G I D + R++++ TLG G+FG+V + +D
Sbjct 10 DAEGHLIYHVGDWLQERYEIVSTLGEGTFGRVVQCVD 46
> CE04173
Length=531
Score = 32.3 bits (72), Expect = 0.21, Method: Composition-based stats.
Identities = 16/56 (28%), Positives = 29/56 (51%), Gaps = 0/56 (0%)
Query 9 SSNRQLMNATSNSACSYFCDADGDYIVSQNDHICYRFQLLQTLGTGSFGQVFRALD 64
S N + + A N+ F + Y + +++HI YR+ + + +G G +G V A D
Sbjct 159 SMNPETLKAPENAIEKDFFEMGSIYKMKKDEHIGYRYTIDKVIGQGCYGVVGEATD 214
> Hs4758008
Length=484
Score = 32.3 bits (72), Expect = 0.23, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 28 DADGDYIVSQNDHICYRFQLLQTLGTGSFGQVFRALD 64
D +G I D + R++++ TLG G+FG+V +D
Sbjct 144 DEEGHLICQSGDVLSARYEIVDTLGEGAFGKVVECID 180
> Hs22045050
Length=484
Score = 32.3 bits (72), Expect = 0.23, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 28 DADGDYIVSQNDHICYRFQLLQTLGTGSFGQVFRALD 64
D +G I D + R++++ TLG G+FG+V +D
Sbjct 144 DEEGHLICQSGDVLSARYEIVDTLGEGAFGKVVECID 180
> SPAC1E11.03
Length=534
Score = 32.0 bits (71), Expect = 0.27, Method: Composition-based stats.
Identities = 23/70 (32%), Positives = 34/70 (48%), Gaps = 6/70 (8%)
Query 2 PRRQRMSSSN--RQLMNATSNSACSYFCDADGDYIVSQNDHICY----RFQLLQTLGTGS 55
P+ Q S N R L + F + + DY++ ND + ++ +L TLG G+
Sbjct 81 PKFQFSSEQNPRRPLTKPSEGVHNHGFDNVNHDYVLYVNDLLGTDEGRKYLILDTLGHGT 140
Query 56 FGQVFRALDL 65
FGQV R DL
Sbjct 141 FGQVARCQDL 150
> YKL166c
Length=398
Score = 31.6 bits (70), Expect = 0.34, Method: Composition-based stats.
Identities = 16/32 (50%), Positives = 20/32 (62%), Gaps = 7/32 (21%)
Query 28 DADGDYIVSQNDHICYRFQLLQTLGTGSFGQV 59
D G Y +S FQ+L+TLGTGSFG+V
Sbjct 78 DTSGKYSLSD-------FQILRTLGTGSFGRV 102
> CE04430
Length=821
Score = 31.2 bits (69), Expect = 0.43, Method: Composition-based stats.
Identities = 19/70 (27%), Positives = 40/70 (57%), Gaps = 9/70 (12%)
Query 2 PRRQRMSSSNRQL----MNATSNSACSY-FCDADGDYIVSQNDHICY----RFQLLQTLG 52
P +SS+N ++ + AT+ + + +G+Y + +N+ +C ++++L+ LG
Sbjct 70 PAAPAISSANNKMAPQSVTATAKTTTNRGKVSGEGEYQLIKNEVLCSPYGNQYEVLEFLG 129
Query 53 TGSFGQVFRA 62
G+FGQV +A
Sbjct 130 KGTFGQVVKA 139
> CE16557
Length=318
Score = 31.2 bits (69), Expect = 0.45, Method: Composition-based stats.
Identities = 11/21 (52%), Positives = 17/21 (80%), Gaps = 0/21 (0%)
Query 45 FQLLQTLGTGSFGQVFRALDL 65
FQ+ + +G G FGQ+FRA+D+
Sbjct 38 FQVEKMVGGGGFGQIFRAVDM 58
> HsM4759010
Length=819
Score = 31.2 bits (69), Expect = 0.50, Method: Composition-based stats.
Identities = 11/21 (52%), Positives = 17/21 (80%), Gaps = 0/21 (0%)
Query 44 RFQLLQTLGTGSFGQVFRALD 64
RF+LLQ +G G++G V++A D
Sbjct 14 RFELLQRVGAGTYGDVYKARD 34
> Hs22035600
Length=820
Score = 31.2 bits (69), Expect = 0.50, Method: Composition-based stats.
Identities = 11/21 (52%), Positives = 17/21 (80%), Gaps = 0/21 (0%)
Query 44 RFQLLQTLGTGSFGQVFRALD 64
RF+LLQ +G G++G V++A D
Sbjct 15 RFELLQRVGAGTYGDVYKARD 35
> YJL164c
Length=397
Score = 31.2 bits (69), Expect = 0.53, Method: Composition-based stats.
Identities = 14/20 (70%), Positives = 18/20 (90%), Gaps = 1/20 (5%)
Query 41 ICYR-FQLLQTLGTGSFGQV 59
I Y+ FQ+L+TLGTGSFG+V
Sbjct 82 IVYKNFQILRTLGTGSFGRV 101
> YLL019c
Length=737
Score = 30.8 bits (68), Expect = 0.56, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 22/39 (56%), Gaps = 2/39 (5%)
Query 28 DADGDYIVSQNDHICY--RFQLLQTLGTGSFGQVFRALD 64
D DG Y+ +ND RF + LG G+FG+V + +D
Sbjct 294 DKDGHYVYQENDIFGSGGRFVVKDLLGQGTFGKVLKCID 332
> Hs10190706
Length=481
Score = 30.8 bits (68), Expect = 0.60, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 28 DADGDYIVSQNDHICYRFQLLQTLGTGSFGQVFRALD 64
D +G I D + R++++ TLG G+FG+V +D
Sbjct 142 DEEGHLICQSGDVLRARYEIVDTLGEGAFGKVVECID 178
> SPAC2C4.14c
Length=312
Score = 30.8 bits (68), Expect = 0.70, Method: Composition-based stats.
Identities = 12/23 (52%), Positives = 18/23 (78%), Gaps = 0/23 (0%)
Query 43 YRFQLLQTLGTGSFGQVFRALDL 65
++++ LQ +G GSFG VFRA D+
Sbjct 4 HQYRDLQLIGQGSFGSVFRAQDV 26
> 7291976
Length=1320
Score = 30.8 bits (68), Expect = 0.70, Method: Composition-based stats.
Identities = 14/36 (38%), Positives = 24/36 (66%), Gaps = 3/36 (8%)
Query 29 ADGDYIVSQND---HICYRFQLLQTLGTGSFGQVFR 61
ADGDY + Q++ + +++L+ LG G+FGQV +
Sbjct 173 ADGDYQLVQHEVLYSLSAEYEVLEFLGRGTFGQVVK 208
> Hs13430860
Length=1198
Score = 30.4 bits (67), Expect = 0.76, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 26/37 (70%), Gaps = 3/37 (8%)
Query 28 DADGDYIVSQNDHICY---RFQLLQTLGTGSFGQVFR 61
+++GDY + Q++ +C +++L+ LG G+FGQV +
Sbjct 179 NSEGDYQLVQHEVLCSMTNTYEVLEFLGRGTFGQVVK 215
> CE28238
Length=1040
Score = 30.0 bits (66), Expect = 0.96, Method: Composition-based stats.
Identities = 19/62 (30%), Positives = 32/62 (51%), Gaps = 13/62 (20%)
Query 1 RPRRQRMSSSNRQLMNATSNSACSYFCDADGDYIVSQNDHICYRFQLLQTLGTGSFGQVF 60
R +R+RM+S N L Y D+D + V ++ + L+ LG G+FG+V+
Sbjct 609 RKQRKRMNSENTVL--------SEYEVDSDPVWEVERS-----KLSLVHMLGEGAFGEVW 655
Query 61 RA 62
+A
Sbjct 656 KA 657
> CE18917
Length=278
Score = 30.0 bits (66), Expect = 0.96, Method: Composition-based stats.
Identities = 12/30 (40%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 35 VSQNDHICYRFQLLQTLGTGSFGQVFRALD 64
++QN I R+ L++ +G G FG V+R +D
Sbjct 5 LAQNRLIHKRYTLVRKIGNGQFGAVWRVID 34
> CE29049
Length=1128
Score = 30.0 bits (66), Expect = 1.0, Method: Composition-based stats.
Identities = 19/62 (30%), Positives = 32/62 (51%), Gaps = 13/62 (20%)
Query 1 RPRRQRMSSSNRQLMNATSNSACSYFCDADGDYIVSQNDHICYRFQLLQTLGTGSFGQVF 60
R +R+RM+S N L Y D+D + V ++ + L+ LG G+FG+V+
Sbjct 609 RKQRKRMNSENTVL--------SEYEVDSDPVWEVERS-----KLSLVHMLGEGAFGEVW 655
Query 61 RA 62
+A
Sbjct 656 KA 657
> ECU11g0180
Length=405
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 20/35 (57%), Gaps = 3/35 (8%)
Query 29 ADGDYIVSQNDHICYRFQLLQTLGTGSFGQVFRAL 63
A GDY+ ++ ++ LGTG+FGQV R +
Sbjct 27 ARGDYLAKNKSQ---KYMIIDMLGTGTFGQVVRCV 58
> CE29048
Length=1126
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 19/62 (30%), Positives = 32/62 (51%), Gaps = 13/62 (20%)
Query 1 RPRRQRMSSSNRQLMNATSNSACSYFCDADGDYIVSQNDHICYRFQLLQTLGTGSFGQVF 60
R +R+RM+S N L Y D+D + V ++ + L+ LG G+FG+V+
Sbjct 609 RKQRKRMNSENTVL--------SEYEVDSDPVWEVERS-----KLSLVHMLGEGAFGEVW 655
Query 61 RA 62
+A
Sbjct 656 KA 657
> CE07599
Length=829
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 9/21 (42%), Positives = 19/21 (90%), Gaps = 0/21 (0%)
Query 45 FQLLQTLGTGSFGQVFRALDL 65
++LLQ +G+G++G+V++A D+
Sbjct 15 YELLQRVGSGTYGEVYKARDI 35
> Hs18594815
Length=609
Score = 29.6 bits (65), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 12/19 (63%), Positives = 14/19 (73%), Gaps = 0/19 (0%)
Query 44 RFQLLQTLGTGSFGQVFRA 62
QLL+ +GTGSFG VFR
Sbjct 312 EVQLLKRIGTGSFGTVFRG 330
> Hs7662170
Length=661
Score = 29.6 bits (65), Expect = 1.6, Method: Composition-based stats.
Identities = 11/24 (45%), Positives = 19/24 (79%), Gaps = 0/24 (0%)
Query 43 YRFQLLQTLGTGSFGQVFRALDLF 66
+R++L +TLG G++G+V RA + F
Sbjct 53 HRYELQETLGKGTYGKVKRATERF 76
> Hs6005810
Length=833
Score = 29.6 bits (65), Expect = 1.6, Method: Composition-based stats.
Identities = 11/20 (55%), Positives = 16/20 (80%), Gaps = 0/20 (0%)
Query 45 FQLLQTLGTGSFGQVFRALD 64
+ LLQ LG G++G+VF+A D
Sbjct 17 YDLLQRLGGGTYGEVFKARD 36
> Hs4502193
Length=606
Score = 29.3 bits (64), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 12/19 (63%), Positives = 14/19 (73%), Gaps = 0/19 (0%)
Query 44 RFQLLQTLGTGSFGQVFRA 62
QLL+ +GTGSFG VFR
Sbjct 309 EVQLLKRIGTGSFGTVFRG 327
> Hs15619015
Length=351
Score = 29.3 bits (64), Expect = 1.6, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 24/44 (54%), Gaps = 5/44 (11%)
Query 21 SACSYFCDADGDYIV-----SQNDHICYRFQLLQTLGTGSFGQV 59
S + A GD++ +QN +F+ L+TLG GSFG+V
Sbjct 15 SVNEFLAKARGDFLYRWGNPAQNTASSDQFERLRTLGMGSFGRV 58
> CE07737
Length=1418
Score = 29.3 bits (64), Expect = 1.8, Method: Composition-based stats.
Identities = 17/63 (26%), Positives = 31/63 (49%), Gaps = 3/63 (4%)
Query 3 RRQRMSSSNRQLMNATSNSACSYFCDADGDYIVSQNDHICYRFQLLQTLGTGSFGQVFRA 62
+R R+ N Q+ ++ F A I ++ +++ LL + +GSFG V RA
Sbjct 1086 KRNRIDYDNLQIGKVITDDNRRNFVLATDKKITTKAP---FQWALLDHIASGSFGSVHRA 1142
Query 63 LDL 65
+D+
Sbjct 1143 MDI 1145
> Hs7657526
Length=745
Score = 29.3 bits (64), Expect = 1.8, Method: Composition-based stats.
Identities = 11/17 (64%), Positives = 15/17 (88%), Gaps = 0/17 (0%)
Query 44 RFQLLQTLGTGSFGQVF 60
+F+LL+ LG GSFG+VF
Sbjct 72 QFELLKVLGQGSFGKVF 88
Lambda K H
0.327 0.136 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1163608362
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40