bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_7973_orf1
Length=81
Score E
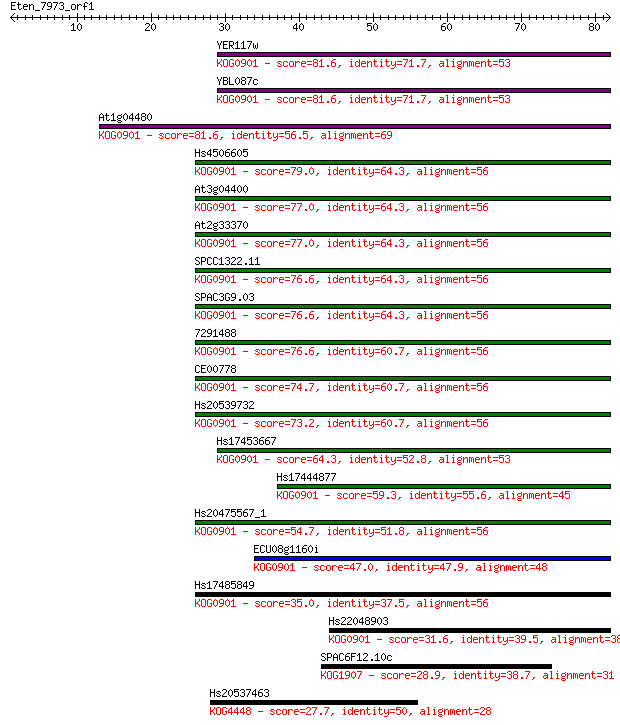
Sequences producing significant alignments: (Bits) Value
YER117w 81.6 3e-16
YBL087c 81.6 3e-16
At1g04480 81.6 3e-16
Hs4506605 79.0 2e-15
At3g04400 77.0 7e-15
At2g33370 77.0 7e-15
SPCC1322.11 76.6 1e-14
SPAC3G9.03 76.6 1e-14
7291488 76.6 1e-14
CE00778 74.7 4e-14
Hs20539732 73.2 1e-13
Hs17453667 64.3 6e-11
Hs17444877 59.3 2e-09
Hs20475567_1 54.7 4e-08
ECU08g1160i 47.0 8e-06
Hs17485849 35.0 0.034
Hs22048903 31.6 0.35
SPAC6F12.10c 28.9 2.2
Hs20537463 27.7 5.3
> YER117w
Length=137
Score = 81.6 bits (200), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 38/53 (71%), Positives = 47/53 (88%), Gaps = 0/53 (0%)
Query 29 GAGGNKMKVTLGLPVGALINCCDNSGAKNLYIIAVKGWGSCLNRLPSSSLGDL 81
GA G K +++LGLPVGA++NC DNSGA+NLYIIAVKG GS LNRLP++SLGD+
Sbjct 5 GAQGTKFRISLGLPVGAIMNCADNSGARNLYIIAVKGSGSRLNRLPAASLGDM 57
> YBL087c
Length=137
Score = 81.6 bits (200), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 38/53 (71%), Positives = 47/53 (88%), Gaps = 0/53 (0%)
Query 29 GAGGNKMKVTLGLPVGALINCCDNSGAKNLYIIAVKGWGSCLNRLPSSSLGDL 81
GA G K +++LGLPVGA++NC DNSGA+NLYIIAVKG GS LNRLP++SLGD+
Sbjct 5 GAQGTKFRISLGLPVGAIMNCADNSGARNLYIIAVKGSGSRLNRLPAASLGDM 57
> At1g04480
Length=175
Score = 81.6 bits (200), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 39/69 (56%), Positives = 52/69 (75%), Gaps = 0/69 (0%)
Query 13 FMLSLLCVAFVCLGRGGAGGNKMKVTLGLPVGALINCCDNSGAKNLYIIAVKGWGSCLNR 72
+L ++ + F LGRGG GNK +++LGLPV A +NC DN+GAKNLYII+VKG LNR
Sbjct 27 LVLIVVDLDFFRLGRGGTSGNKFRMSLGLPVAATVNCADNTGAKNLYIISVKGIKGRLNR 86
Query 73 LPSSSLGDL 81
LPS+ +GD+
Sbjct 87 LPSACVGDM 95
> Hs4506605
Length=140
Score = 79.0 bits (193), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 36/56 (64%), Positives = 47/56 (83%), Gaps = 0/56 (0%)
Query 26 GRGGAGGNKMKVTLGLPVGALINCCDNSGAKNLYIIAVKGWGSCLNRLPSSSLGDL 81
GRGG+ G K +++LGLPVGA+INC DN+GAKNLYII+VKG LNRLP++ +GD+
Sbjct 5 GRGGSSGAKFRISLGLPVGAVINCADNTGAKNLYIISVKGIKGRLNRLPAAGVGDM 60
> At3g04400
Length=140
Score = 77.0 bits (188), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 36/56 (64%), Positives = 45/56 (80%), Gaps = 0/56 (0%)
Query 26 GRGGAGGNKMKVTLGLPVGALINCCDNSGAKNLYIIAVKGWGSCLNRLPSSSLGDL 81
GRGG GNK +++LGLPV A +NC DN+GAKNLYII+VKG LNRLPS+ +GD+
Sbjct 5 GRGGTSGNKFRMSLGLPVAATVNCADNTGAKNLYIISVKGIKGRLNRLPSACVGDM 60
> At2g33370
Length=140
Score = 77.0 bits (188), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 36/56 (64%), Positives = 45/56 (80%), Gaps = 0/56 (0%)
Query 26 GRGGAGGNKMKVTLGLPVGALINCCDNSGAKNLYIIAVKGWGSCLNRLPSSSLGDL 81
GRGG GNK +++LGLPV A +NC DN+GAKNLYII+VKG LNRLPS+ +GD+
Sbjct 5 GRGGTSGNKFRMSLGLPVAATVNCADNTGAKNLYIISVKGIKGRLNRLPSACVGDM 60
> SPCC1322.11
Length=139
Score = 76.6 bits (187), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 36/56 (64%), Positives = 46/56 (82%), Gaps = 0/56 (0%)
Query 26 GRGGAGGNKMKVTLGLPVGALINCCDNSGAKNLYIIAVKGWGSCLNRLPSSSLGDL 81
GRG A G K ++TLGLPV A++NC DNSGAKNLYI++V G G+ LNRLP++S GD+
Sbjct 4 GRGAASGTKYRMTLGLPVQAIMNCADNSGAKNLYIVSVFGTGARLNRLPAASCGDM 59
> SPAC3G9.03
Length=139
Score = 76.6 bits (187), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 36/56 (64%), Positives = 46/56 (82%), Gaps = 0/56 (0%)
Query 26 GRGGAGGNKMKVTLGLPVGALINCCDNSGAKNLYIIAVKGWGSCLNRLPSSSLGDL 81
GRG A G K ++TLGLPV A++NC DNSGAKNLYI++V G G+ LNRLP++S GD+
Sbjct 4 GRGAASGTKYRMTLGLPVQAIMNCADNSGAKNLYIVSVFGTGARLNRLPAASCGDM 59
> 7291488
Length=140
Score = 76.6 bits (187), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 34/56 (60%), Positives = 45/56 (80%), Gaps = 0/56 (0%)
Query 26 GRGGAGGNKMKVTLGLPVGALINCCDNSGAKNLYIIAVKGWGSCLNRLPSSSLGDL 81
GRGG G K +++LGLPVGA++NC DN+GAKNLY+IAV G LNRLP++ +GD+
Sbjct 5 GRGGTAGGKFRISLGLPVGAVMNCADNTGAKNLYVIAVHGIRGRLNRLPAAGVGDM 60
> CE00778
Length=140
Score = 74.7 bits (182), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 34/56 (60%), Positives = 46/56 (82%), Gaps = 0/56 (0%)
Query 26 GRGGAGGNKMKVTLGLPVGALINCCDNSGAKNLYIIAVKGWGSCLNRLPSSSLGDL 81
GRGGA G K +++LGLPVGA++NC DN+GAKNL++I+V G LNRLPS+ +GD+
Sbjct 5 GRGGASGAKFRISLGLPVGAVMNCADNTGAKNLFVISVYGIRGRLNRLPSAGVGDM 60
> Hs20539732
Length=107
Score = 73.2 bits (178), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 34/56 (60%), Positives = 46/56 (82%), Gaps = 0/56 (0%)
Query 26 GRGGAGGNKMKVTLGLPVGALINCCDNSGAKNLYIIAVKGWGSCLNRLPSSSLGDL 81
GRGG+ G ++++GLPVGA+INC DN+GAKNLYII+VKG LNRLP++ +GD+
Sbjct 5 GRGGSSGVNFRISVGLPVGAVINCADNTGAKNLYIISVKGIKGRLNRLPAAGVGDM 60
> Hs17453667
Length=243
Score = 64.3 bits (155), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 28/53 (52%), Positives = 41/53 (77%), Gaps = 0/53 (0%)
Query 29 GAGGNKMKVTLGLPVGALINCCDNSGAKNLYIIAVKGWGSCLNRLPSSSLGDL 81
G+ G K +++LGLP+G +INC N+ AKNLY+I+VKG LNRLP++ +GD+
Sbjct 95 GSLGVKFRISLGLPIGTMINCAGNTEAKNLYVISVKGIKGELNRLPAAGMGDM 147
> Hs17444877
Length=113
Score = 59.3 bits (142), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 25/45 (55%), Positives = 35/45 (77%), Gaps = 0/45 (0%)
Query 37 VTLGLPVGALINCCDNSGAKNLYIIAVKGWGSCLNRLPSSSLGDL 81
++L PVGA +NC DN+GAK+LYII+ KG LNRLP++ +GD+
Sbjct 1 MSLSFPVGAAVNCADNTGAKSLYIISTKGIKGRLNRLPAADVGDM 45
> Hs20475567_1
Length=106
Score = 54.7 bits (130), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 29/56 (51%), Positives = 38/56 (67%), Gaps = 0/56 (0%)
Query 26 GRGGAGGNKMKVTLGLPVGALINCCDNSGAKNLYIIAVKGWGSCLNRLPSSSLGDL 81
G GG K +LG +GA INC DN+GAKNLYI++VKG LNRL ++ +GD+
Sbjct 5 GCGGFSSVKYWNSLGFLIGAKINCVDNTGAKNLYIMSVKGIKGQLNRLSTAGVGDM 60
> ECU08g1160i
Length=146
Score = 47.0 bits (110), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 23/48 (47%), Positives = 32/48 (66%), Gaps = 0/48 (0%)
Query 34 KMKVTLGLPVGALINCCDNSGAKNLYIIAVKGWGSCLNRLPSSSLGDL 81
+ K+T G+ V L+ C DNSGAK L I VK + LNRLP+++ GD+
Sbjct 19 RYKMTRGIQVETLMKCADNSGAKILRCIGVKRYRGRLNRLPAAAPGDI 66
> Hs17485849
Length=200
Score = 35.0 bits (79), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 30/56 (53%), Gaps = 0/56 (0%)
Query 26 GRGGAGGNKMKVTLGLPVGALINCCDNSGAKNLYIIAVKGWGSCLNRLPSSSLGDL 81
GR G+ K ++LGL +G KNL I +VKG LNRL ++ +GD+
Sbjct 79 GRDGSSSMKFWISLGLLLGVESTVLTTQELKNLCIFSVKGIKGWLNRLSAAGVGDV 134
> Hs22048903
Length=325
Score = 31.6 bits (70), Expect = 0.35, Method: Composition-based stats.
Identities = 15/38 (39%), Positives = 24/38 (63%), Gaps = 0/38 (0%)
Query 44 GALINCCDNSGAKNLYIIAVKGWGSCLNRLPSSSLGDL 81
G +I+ D + A+NL+II +K LNRLP + +G +
Sbjct 27 GPVIHYADGTEAENLHIIVMKEIKRQLNRLPDAGVGAM 64
> SPAC6F12.10c
Length=1323
Score = 28.9 bits (63), Expect = 2.2, Method: Composition-based stats.
Identities = 12/31 (38%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 43 VGALINCCDNSGAKNLYIIAVKGWGSCLNRL 73
+GA+I CD AK L + A G +C++R+
Sbjct 944 LGAVIQVCDRDIAKVLELFAANGLSTCVHRI 974
> Hs20537463
Length=339
Score = 27.7 bits (60), Expect = 5.3, Method: Composition-based stats.
Identities = 14/28 (50%), Positives = 17/28 (60%), Gaps = 1/28 (3%)
Query 28 GGAGGNKMKVTLGLPVGALINCCDNSGA 55
G GG KMK+TLG P G + C+ S A
Sbjct 166 GPGGGTKMKLTLG-PHGIRMQPCERSAA 192
Lambda K H
0.323 0.142 0.453
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1162440328
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40