bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_8010_orf1
Length=70
Score E
Sequences producing significant alignments: (Bits) Value
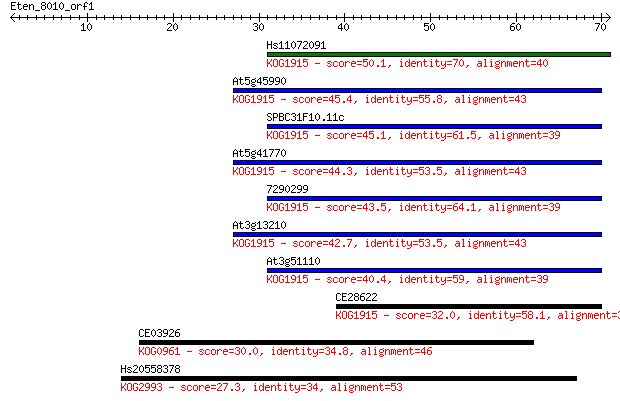
Hs11072091 50.1 9e-07
At5g45990 45.4 2e-05
SPBC31F10.11c 45.1 3e-05
At5g41770 44.3 6e-05
7290299 43.5 9e-05
At3g13210 42.7 2e-04
At3g51110 40.4 8e-04
CE28622 32.0 0.27
CE03926 30.0 1.2
Hs20558378 27.3 7.8
> Hs11072091
Length=836
Score = 50.1 bits (118), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 28/40 (70%), Positives = 30/40 (75%), Gaps = 1/40 (2%)
Query 31 VKNKMPAAVQITAEQLLREAVDRQLEDAAAAKPQQRIVDE 70
VKNK PA VQITAEQLLREA +R+LE PQQRI DE
Sbjct 167 VKNKAPAEVQITAEQLLREAKERELE-LLPPPPQQRITDE 205
> At5g45990
Length=673
Score = 45.4 bits (106), Expect = 2e-05, Method: Composition-based stats.
Identities = 24/43 (55%), Positives = 30/43 (69%), Gaps = 2/43 (4%)
Query 27 KQMEVKNKMPAAVQITAEQLLREAVDRQLEDAAAAKPQQRIVD 69
+ VKNK PA VQITAEQ+LREA +RQ +A P+Q+I D
Sbjct 12 RTTRVKNKTPAPVQITAEQILREARERQ--EAEIRPPKQKITD 52
> SPBC31F10.11c
Length=674
Score = 45.1 bits (105), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 24/39 (61%), Positives = 28/39 (71%), Gaps = 2/39 (5%)
Query 31 VKNKMPAAVQITAEQLLREAVDRQLEDAAAAKPQQRIVD 69
VKNK PA +QI+AEQLLREAV+RQ D A P+ I D
Sbjct 8 VKNKNPAPIQISAEQLLREAVERQ--DVAFVPPKINITD 44
> At5g41770
Length=665
Score = 44.3 bits (103), Expect = 6e-05, Method: Composition-based stats.
Identities = 23/43 (53%), Positives = 30/43 (69%), Gaps = 2/43 (4%)
Query 27 KQMEVKNKMPAAVQITAEQLLREAVDRQLEDAAAAKPQQRIVD 69
+ VKNK PA +QITAEQ+LREA +RQ +A P+Q+I D
Sbjct 12 RPTRVKNKTPAPIQITAEQILREARERQ--EAEIRPPKQKITD 52
> 7290299
Length=702
Score = 43.5 bits (101), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 25/39 (64%), Positives = 28/39 (71%), Gaps = 1/39 (2%)
Query 31 VKNKMPAAVQITAEQLLREAVDRQLEDAAAAKPQQRIVD 69
VKNK PA VQITAEQLLREA +R LE P+Q+I D
Sbjct 13 VKNKAPAEVQITAEQLLREAKERDLE-ILPPPPKQKISD 50
> At3g13210
Length=657
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 23/43 (53%), Positives = 29/43 (67%), Gaps = 2/43 (4%)
Query 27 KQMEVKNKMPAAVQITAEQLLREAVDRQLEDAAAAKPQQRIVD 69
+ +VKNK PA +QITAEQ+LREA +RQ +A P Q I D
Sbjct 25 RMTQVKNKTPAPIQITAEQILREARERQ--EAEFRPPNQTITD 65
> At3g51110
Length=599
Score = 40.4 bits (93), Expect = 8e-04, Method: Composition-based stats.
Identities = 23/40 (57%), Positives = 30/40 (75%), Gaps = 3/40 (7%)
Query 31 VKNKMPAAVQITAEQLLREAVDRQLEDAAAAK-PQQRIVD 69
VKNK PA VQITAEQ+L+EA R+ ED+ + P+Q+I D
Sbjct 2 VKNKTPAPVQITAEQVLKEA--REREDSRILRPPKQKITD 39
> CE28622
Length=744
Score = 32.0 bits (71), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 18/31 (58%), Positives = 23/31 (74%), Gaps = 1/31 (3%)
Query 39 VQITAEQLLREAVDRQLEDAAAAKPQQRIVD 69
+QITAEQLLREA +R+LE A P+ +I D
Sbjct 31 LQITAEQLLREAKERELELIPPA-PKTKITD 60
> CE03926
Length=995
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 25/48 (52%), Gaps = 2/48 (4%)
Query 16 GAKNYPLGASWKQMEVKNKM--PAAVQITAEQLLREAVDRQLEDAAAA 61
GA YPL A W Q+ + + P+ + A++L EA DR+ + A
Sbjct 633 GADKYPLLAKWAQIFTQGVVFDPSRIHQCAQKLAGEARDRKRDGCTVA 680
> Hs20558378
Length=1938
Score = 27.3 bits (59), Expect = 7.8, Method: Composition-based stats.
Identities = 18/58 (31%), Positives = 32/58 (55%), Gaps = 5/58 (8%)
Query 14 RRGAKNYPLGASWKQMEVKNKMPAAVQITAE---QLLREA--VDRQLEDAAAAKPQQR 66
+RGA ++ + +E+ N++ A+Q TAE +L A V R L+D +A+ +R
Sbjct 1783 QRGAASFGSSTASAALELSNRLVQAIQATAETVYDILSPAAPVSRSLQDKRSARRLRR 1840
Lambda K H
0.313 0.130 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1197219688
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40