bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_8011_orf1
Length=79
Score E
Sequences producing significant alignments: (Bits) Value
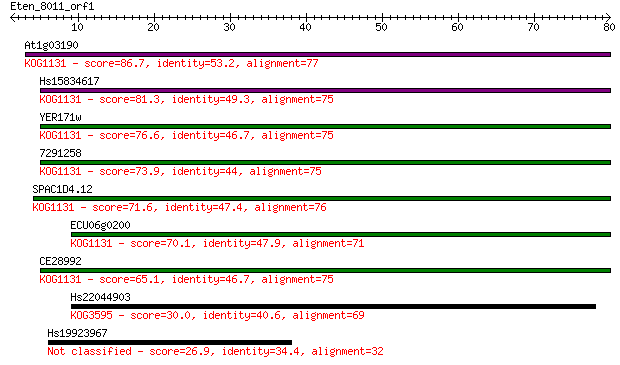
At1g03190 86.7 9e-18
Hs15834617 81.3 4e-16
YER171w 76.6 9e-15
7291258 73.9 7e-14
SPAC1D4.12 71.6 4e-13
ECU06g0200 70.1 8e-13
CE28992 65.1 3e-11
Hs22044903 30.0 1.1
Hs19923967 26.9 8.3
> At1g03190
Length=758
Score = 86.7 bits (213), Expect = 9e-18, Method: Composition-based stats.
Identities = 41/78 (52%), Positives = 56/78 (71%), Gaps = 1/78 (1%)
Query 3 LALQVTDLERLLPLTLVADFCTLVGTYWDGFIVIVDPYPE-ASGLHDPLLQLCCLDASLA 61
L L++TD + LP+ V DF TLVGTY GF +I++PY E + DP+LQL C DASLA
Sbjct 384 LTLEITDTDEFLPIQTVCDFATLVGTYARGFSIIIEPYDERMPHIPDPILQLSCHDASLA 443
Query 62 MQQVLSRFKSVILTSGTI 79
++ V RF+SV++TSGT+
Sbjct 444 IKPVFDRFQSVVITSGTL 461
> Hs15834617
Length=760
Score = 81.3 bits (199), Expect = 4e-16, Method: Composition-based stats.
Identities = 37/76 (48%), Positives = 56/76 (73%), Gaps = 1/76 (1%)
Query 5 LQVTDLERLLPLTLVADFCTLVGTYWDGFIVIVDPYPEAS-GLHDPLLQLCCLDASLAMQ 63
L++TDL PLTL+A+F TLV TY GF +I++P+ + + + +P+L C+DASLA++
Sbjct 386 LEITDLADFSPLTLLANFATLVSTYAKGFTIIIEPFDDRTPTIANPILHFSCMDASLAIK 445
Query 64 QVLSRFKSVILTSGTI 79
V RF+SVI+TSGT+
Sbjct 446 PVFERFQSVIITSGTL 461
> YER171w
Length=778
Score = 76.6 bits (187), Expect = 9e-15, Method: Composition-based stats.
Identities = 35/76 (46%), Positives = 55/76 (72%), Gaps = 1/76 (1%)
Query 5 LQVTDLERLLPLTLVADFCTLVGTYWDGFIVIVDPYP-EASGLHDPLLQLCCLDASLAMQ 63
L+VT++E L +A F TL+ TY +GF++I++PY E + + +P+++ CLDAS+A++
Sbjct 388 LEVTEVEDFTALKDIATFATLISTYEEGFLLIIEPYEIENAAVPNPIMRFTCLDASIAIK 447
Query 64 QVLSRFKSVILTSGTI 79
V RF SVI+TSGTI
Sbjct 448 PVFERFSSVIITSGTI 463
> 7291258
Length=745
Score = 73.9 bits (180), Expect = 7e-14, Method: Composition-based stats.
Identities = 33/76 (43%), Positives = 53/76 (69%), Gaps = 1/76 (1%)
Query 5 LQVTDLERLLPLTLVADFCTLVGTYWDGFIVIVDPYPEAS-GLHDPLLQLCCLDASLAMQ 63
L+++DL LTL+ F TLV TY GF +I++P+ + + + +P+L CLD+S+AM
Sbjct 362 LEISDLTEYGALTLITHFATLVSTYTKGFTIIIEPFDDKTPTVSNPILHFSCLDSSIAMA 421
Query 64 QVLSRFKSVILTSGTI 79
V SRF++V++TSGT+
Sbjct 422 PVFSRFQTVVITSGTL 437
> SPAC1D4.12
Length=772
Score = 71.6 bits (174), Expect = 4e-13, Method: Composition-based stats.
Identities = 36/77 (46%), Positives = 54/77 (70%), Gaps = 1/77 (1%)
Query 4 ALQVTDLERLLPLTLVADFCTLVGTYWDGFIVIVDPY-PEASGLHDPLLQLCCLDASLAM 62
ALQ++ +E L V F TLV TY GFI+I++P+ E + + +P+L+ CLDAS+A+
Sbjct 384 ALQISLVEDFHSLQQVVAFATLVATYERGFILILEPFETENATVPNPILRFSCLDASIAI 443
Query 63 QQVLSRFKSVILTSGTI 79
+ V RF+SVI+TSGT+
Sbjct 444 KPVFERFRSVIITSGTL 460
> ECU06g0200
Length=742
Score = 70.1 bits (170), Expect = 8e-13, Method: Composition-based stats.
Identities = 34/72 (47%), Positives = 51/72 (70%), Gaps = 1/72 (1%)
Query 9 DLERLLPLTLVADFCTLVGTYWDGFIVIVDPY-PEASGLHDPLLQLCCLDASLAMQQVLS 67
D E + L VADF T+V Y GF+VI +P+ +AS + +P L+L CLD+S+A+ V +
Sbjct 367 DDEDMGHLKTVADFSTMVSMYSKGFVVIFEPFDSQASTVFNPTLRLACLDSSIAISPVFA 426
Query 68 RFKSVILTSGTI 79
RF++VI+TSGT+
Sbjct 427 RFRNVIITSGTM 438
> CE28992
Length=751
Score = 65.1 bits (157), Expect = 3e-11, Method: Composition-based stats.
Identities = 35/75 (46%), Positives = 49/75 (65%), Gaps = 3/75 (4%)
Query 5 LQVTDLERLLPLTLVADFCTLVGTYWDGFIVIVDPYPEASGLHDPLLQLCCLDASLAMQQ 64
L++TD + L+ V CTLV TY GF VIV+P G ++ L C DAS+A++
Sbjct 380 LEITDNGDVWALSQVTTLCTLVSTYSKGFSVIVEP---QDGSQLAVITLSCHDASIAIRP 436
Query 65 VLSRFKSVILTSGTI 79
V+SRF+SVI+TSGT+
Sbjct 437 VMSRFQSVIITSGTL 451
> Hs22044903
Length=2252
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 28/93 (30%), Positives = 39/93 (41%), Gaps = 30/93 (32%)
Query 9 DLERLLPLTLVADFCTLV--------------------GTYWDGFIVI----VDPYPEAS 44
D +R L LT++ADF L GTY D I +PE
Sbjct 2106 DWDRRLLLTMLADFYNLYIVENPHYKFSPSGNYFAPPKGTYEDYIEFIKKLPFTQHPEIF 2165
Query 45 GLHDPLLQLCCLDASLAMQQVLSRFKSVILTSG 77
GLH+ + D S +QQ + F+S++LT G
Sbjct 2166 GLHENV------DISKDLQQTKTLFESLLLTQG 2192
> Hs19923967
Length=838
Score = 26.9 bits (58), Expect = 8.3, Method: Composition-based stats.
Identities = 11/32 (34%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 6 QVTDLERLLPLTLVADFCTLVGTYWDGFIVIV 37
Q+T + L LT+ D+ TL+ Y GF+ ++
Sbjct 696 QLTGIRMLRHLTMTIDYHTLIANYMSGFLSLL 727
Lambda K H
0.326 0.141 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1168763848
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40