bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_8074_orf1
Length=79
Score E
Sequences producing significant alignments: (Bits) Value
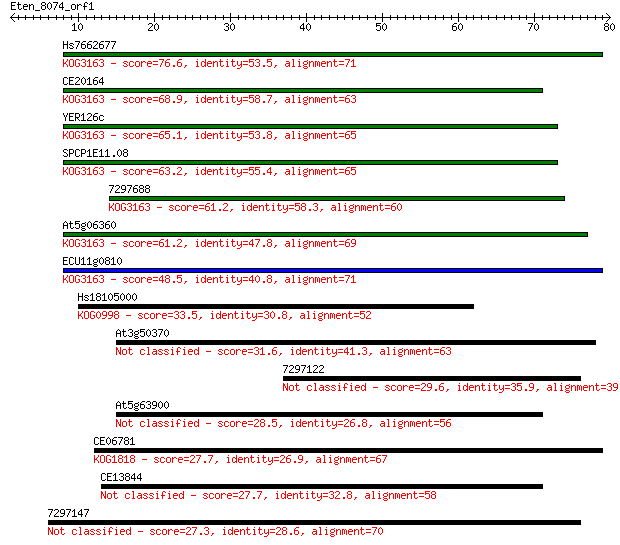
Hs7662677 76.6 9e-15
CE20164 68.9 2e-12
YER126c 65.1 3e-11
SPCP1E11.08 63.2 1e-10
7297688 61.2 4e-10
At5g06360 61.2 5e-10
ECU11g0810 48.5 3e-06
Hs18105000 33.5 0.088
At3g50370 31.6 0.40
7297122 29.6 1.4
At5g63900 28.5 3.2
CE06781 27.7 5.1
CE13844 27.7 5.5
7297147 27.3 6.2
> Hs7662677
Length=260
Score = 76.6 bits (187), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 38/71 (53%), Positives = 57/71 (80%), Gaps = 0/71 (0%)
Query 8 MPQNEYIEQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKRFLEKQK 67
MPQNEYIE H+KR+G R D E++RKKE+RE HER+++AK + G+KAK+ +K+R EK +
Sbjct 1 MPQNEYIELHRKRYGYRLDYHEKKRKKESREAHERSKKAKKMIGLKAKLYHKQRHAEKIQ 60
Query 68 MKKRIQLEDRK 78
MKK I++ +++
Sbjct 61 MKKTIKMHEKR 71
> CE20164
Length=259
Score = 68.9 bits (167), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 37/63 (58%), Positives = 48/63 (76%), Gaps = 0/63 (0%)
Query 8 MPQNEYIEQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKRFLEKQK 67
MPQNE+IE H+KR G R D ER+RKK AR H+R++ AK LRG KAK+ +KKR+ EK +
Sbjct 1 MPQNEHIELHRKRHGRRLDHEERQRKKLARAAHDRSQMAKTLRGHKAKLYHKKRYSEKVE 60
Query 68 MKK 70
M+K
Sbjct 61 MRK 63
> YER126c
Length=261
Score = 65.1 bits (157), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 35/65 (53%), Positives = 47/65 (72%), Gaps = 0/65 (0%)
Query 8 MPQNEYIEQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKRFLEKQK 67
MPQN+YIE+H K+ G+R D ER+RK+EARE H+ + RA+ L G K K KKR+ EK
Sbjct 1 MPQNDYIERHIKQHGKRLDHEERKRKREARESHKISERAQKLTGWKGKQFAKKRYAEKVS 60
Query 68 MKKRI 72
M+K+I
Sbjct 61 MRKKI 65
> SPCP1E11.08
Length=260
Score = 63.2 bits (152), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 36/65 (55%), Positives = 46/65 (70%), Gaps = 0/65 (0%)
Query 8 MPQNEYIEQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKRFLEKQK 67
MPQNEYIE+ ++ G RFD ER+RKK ARE H+ + A+ RGIKAK+ +KR EK +
Sbjct 1 MPQNEYIEESIRKHGRRFDHEERKRKKAAREAHDASLYAQKTRGIKAKLYQEKRRKEKIQ 60
Query 68 MKKRI 72
MKK I
Sbjct 61 MKKTI 65
> 7297688
Length=253
Score = 61.2 bits (147), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 35/60 (58%), Positives = 48/60 (80%), Gaps = 0/60 (0%)
Query 14 IEQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKRFLEKQKMKKRIQ 73
+E+H+K +G R D ER+RKKEAR P +RAR+A+ LRGIKAK+ NK+R EK ++KK+IQ
Sbjct 1 MERHRKLYGRRLDYEERKRKKEARLPKDRARKARKLRGIKAKLFNKERRNEKIQIKKKIQ 60
> At5g06360
Length=260
Score = 61.2 bits (147), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 33/69 (47%), Positives = 46/69 (66%), Gaps = 0/69 (0%)
Query 8 MPQNEYIEQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKRFLEKQK 67
MPQ +YI+ H+KR G R D ER+RKKEARE H+ + A+ GIK K+ KK + EK
Sbjct 1 MPQGDYIDLHRKRNGYRLDHFERKRKKEAREVHKHSTMAQKSLGIKGKMIAKKNYAEKAL 60
Query 68 MKKRIQLED 76
MKK +++ +
Sbjct 61 MKKTLKMHE 69
> ECU11g0810
Length=258
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 29/71 (40%), Positives = 47/71 (66%), Gaps = 0/71 (0%)
Query 8 MPQNEYIEQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKRFLEKQK 67
MPQN+++E+H K++G R D R+ KKE R A+ AK L G+KAK+ KK+ K +
Sbjct 1 MPQNDFVERHIKKYGRRLDHELRKYKKERRNEKLVAKTAKRLFGLKAKLFAKKQHAAKIQ 60
Query 68 MKKRIQLEDRK 78
+KK +++++ K
Sbjct 61 LKKDLKMKESK 71
> Hs18105000
Length=689
Score = 33.5 bits (75), Expect = 0.088, Method: Composition-based stats.
Identities = 16/52 (30%), Positives = 29/52 (55%), Gaps = 2/52 (3%)
Query 10 QNEYIEQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKR 61
Q ++ E+ +KRF ++ E RK+ R+ E+ ++ +LL +K K K R
Sbjct 116 QKQFAEEQQKRFEQQQKLLEEERKR--RQFEEQKQKLRLLSSVKPKTGEKSR 165
> At3g50370
Length=2152
Score = 31.6 bits (70), Expect = 0.40, Method: Composition-based stats.
Identities = 26/77 (33%), Positives = 41/77 (53%), Gaps = 16/77 (20%)
Query 15 EQHKKRFGERFDAAERRRKKEAREPH--------ERARRAKLLRGIKAKVENKKR-FLEK 65
E+ + R D +RR ++EARE E RRA+ LR K+K E K R F+E+
Sbjct 502 EEERLRLAREQDERQRRLEEEAREAAFRNEQERLEATRRAEELR--KSKEEEKHRLFMEE 559
Query 66 QKMK-----KRIQLEDR 77
++ K K ++LE++
Sbjct 560 ERRKQAAKQKLLELEEK 576
> 7297122
Length=1009
Score = 29.6 bits (65), Expect = 1.4, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 23/39 (58%), Gaps = 0/39 (0%)
Query 37 REPHERARRAKLLRGIKAKVENKKRFLEKQKMKKRIQLE 75
R P A+R + + +K E +R LE+Q+ KK +QL+
Sbjct 708 RSPTASAKRIQPITVLKKSDEEWQRHLEQQQQKKHVQLQ 746
> At5g63900
Length=557
Score = 28.5 bits (62), Expect = 3.2, Method: Composition-based stats.
Identities = 15/56 (26%), Positives = 29/56 (51%), Gaps = 0/56 (0%)
Query 15 EQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKRFLEKQKMKK 70
+Q K R G D ++KK + E RR K+++ +K ++ ++ +K K +K
Sbjct 185 DQEKIRIGLNVDVIHEQQKKRRKMASEEIRRPKMVKSLKKVLQVMEKKQQKNKHEK 240
> CE06781
Length=765
Score = 27.7 bits (60), Expect = 5.1, Method: Composition-based stats.
Identities = 18/67 (26%), Positives = 32/67 (47%), Gaps = 5/67 (7%)
Query 12 EYIEQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKRFLEKQKMKKR 71
E ++ H GE R+ E RE HER R+ ++ + + ++ LE +MKK
Sbjct 500 ESLQDHLANIGE-----ARQAIDEMREEHERKRQERMAEEQRLRQAQMQQTLEMMRMKKH 554
Query 72 IQLEDRK 78
L +++
Sbjct 555 AMLMEQR 561
> CE13844
Length=908
Score = 27.7 bits (60), Expect = 5.5, Method: Composition-based stats.
Identities = 19/62 (30%), Positives = 34/62 (54%), Gaps = 5/62 (8%)
Query 13 YIEQHKKRFGERFDAAERRRKKEAREPHERARRAKLLR----GIKAKVENKKRFLEKQKM 68
Y+EQH +R +++ E+R K+ +E A+ ++R G+ + E+K L +QKM
Sbjct 439 YMEQHVERLLQQYKEREKRMKQLEKE-MVSAQLPDIMRNKMLGLLQQKESKYTRLRRQKM 497
Query 69 KK 70
K
Sbjct 498 SK 499
> 7297147
Length=1377
Score = 27.3 bits (59), Expect = 6.2, Method: Composition-based stats.
Identities = 20/73 (27%), Positives = 41/73 (56%), Gaps = 3/73 (4%)
Query 6 FKMPQNEYIEQHKKRFGERF--DAAERRRKKEAREPHERARR-AKLLRGIKAKVENKKRF 62
F+ P N Y + +++ +R +A E++R +E + +R + A L + + + E +KR
Sbjct 434 FENPNNYYKQLQEEQEKQRLVAEAEEQKRLQEEADQQKRLQEEAALNKRLLEEAEQQKRL 493
Query 63 LEKQKMKKRIQLE 75
E+ + +KR+Q E
Sbjct 494 QEEAEQQKRLQEE 506
Lambda K H
0.323 0.138 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1168763848
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40