bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_8231_orf1
Length=86
Score E
Sequences producing significant alignments: (Bits) Value
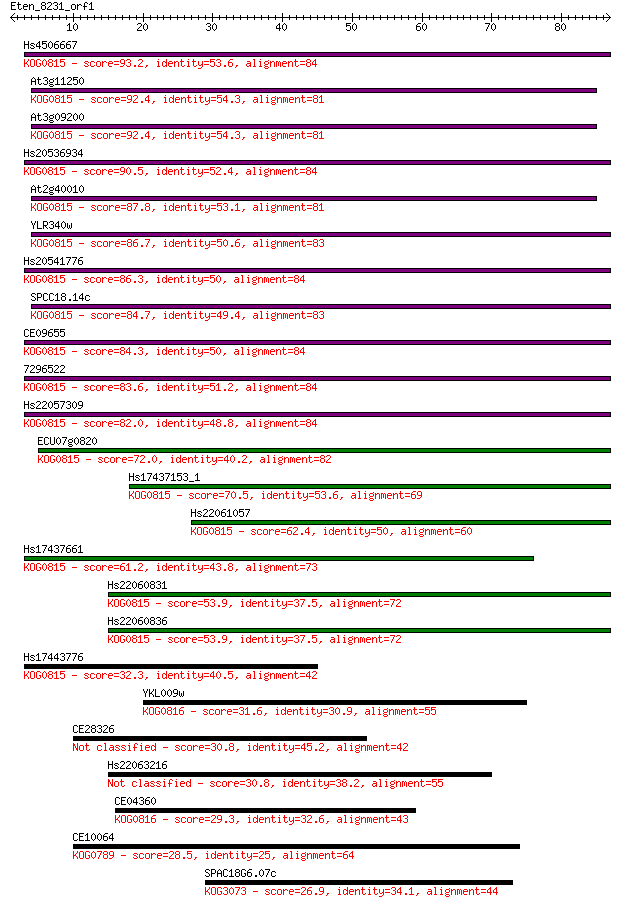
Hs4506667 93.2 9e-20
At3g11250 92.4 2e-19
At3g09200 92.4 2e-19
Hs20536934 90.5 6e-19
At2g40010 87.8 5e-18
YLR340w 86.7 1e-17
Hs20541776 86.3 1e-17
SPCC18.14c 84.7 4e-17
CE09655 84.3 5e-17
7296522 83.6 8e-17
Hs22057309 82.0 3e-16
ECU07g0820 72.0 3e-13
Hs17437153_1 70.5 7e-13
Hs22061057 62.4 2e-10
Hs17437661 61.2 4e-10
Hs22060831 53.9 7e-08
Hs22060836 53.9 8e-08
Hs17443776 32.3 0.20
YKL009w 31.6 0.34
CE28326 30.8 0.60
Hs22063216 30.8 0.73
CE04360 29.3 1.9
CE10064 28.5 2.9
SPAC18G6.07c 26.9 8.5
> Hs4506667
Length=317
Score = 93.2 bits (230), Expect = 9e-20, Method: Composition-based stats.
Identities = 45/84 (53%), Positives = 62/84 (73%), Gaps = 0/84 (0%)
Query 3 RSTQTDPGSTGFFQALGISTKIVKGQIEIQQAVQLIRRGERVSASAATLLHKLSVKPFTY 62
++T P T FFQALGI+TKI +G IEI VQLI+ G++V AS ATLL+ L++ PF++
Sbjct 126 QNTGLGPEKTSFFQALGITTKISRGTIEILSDVQLIKTGDKVGASEATLLNMLNISPFSF 185
Query 63 GLKVEHVYDNGSVYSGWGLDITVE 86
GL ++ V+DNGS+Y+ LDIT E
Sbjct 186 GLVIQQVFDNGSIYNPEVLDITEE 209
> At3g11250
Length=323
Score = 92.4 bits (228), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 44/81 (54%), Positives = 58/81 (71%), Gaps = 0/81 (0%)
Query 4 STQTDPGSTGFFQALGISTKIVKGQIEIQQAVQLIRRGERVSASAATLLHKLSVKPFTYG 63
+T DP T FFQ L I TKI KG +EI V+LI++G++V +S A LL KL ++PF+YG
Sbjct 129 NTGLDPSQTSFFQVLNIPTKINKGTVEIITPVELIKQGDKVGSSEAALLAKLGIRPFSYG 188
Query 64 LKVEHVYDNGSVYSGWGLDIT 84
L V+ VYDNGSV+S LD+T
Sbjct 189 LVVQSVYDNGSVFSPEVLDLT 209
> At3g09200
Length=320
Score = 92.4 bits (228), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 44/81 (54%), Positives = 58/81 (71%), Gaps = 0/81 (0%)
Query 4 STQTDPGSTGFFQALGISTKIVKGQIEIQQAVQLIRRGERVSASAATLLHKLSVKPFTYG 63
+T DP T FFQ L I TKI KG +EI V+LI++G++V +S A LL KL ++PF+YG
Sbjct 129 NTGLDPSQTSFFQVLNIPTKINKGTVEIITPVELIKQGDKVGSSEAALLAKLGIRPFSYG 188
Query 64 LKVEHVYDNGSVYSGWGLDIT 84
L V+ VYDNGSV+S LD+T
Sbjct 189 LVVQSVYDNGSVFSPEVLDLT 209
> Hs20536934
Length=317
Score = 90.5 bits (223), Expect = 6e-19, Method: Composition-based stats.
Identities = 44/84 (52%), Positives = 61/84 (72%), Gaps = 0/84 (0%)
Query 3 RSTQTDPGSTGFFQALGISTKIVKGQIEIQQAVQLIRRGERVSASAATLLHKLSVKPFTY 62
++T P T FFQALGI+TKI +G IEI VQLI+ G++V AS ATLL+ L++ PF++
Sbjct 126 QNTGLGPEKTSFFQALGITTKISRGTIEILSDVQLIKTGDKVGASEATLLNMLNISPFSF 185
Query 63 GLKVEHVYDNGSVYSGWGLDITVE 86
GL ++ V+DNGS+Y+ LD T E
Sbjct 186 GLVIQQVFDNGSIYNPEVLDKTEE 209
> At2g40010
Length=317
Score = 87.8 bits (216), Expect = 5e-18, Method: Composition-based stats.
Identities = 43/81 (53%), Positives = 58/81 (71%), Gaps = 0/81 (0%)
Query 4 STQTDPGSTGFFQALGISTKIVKGQIEIQQAVQLIRRGERVSASAATLLHKLSVKPFTYG 63
+T DP T FFQ L I TKI KG +EI V+LI++G++V +S A LL KL ++PF+YG
Sbjct 130 NTGLDPSQTSFFQVLNIPTKINKGTVEIITPVELIKKGDKVGSSEAALLAKLGIRPFSYG 189
Query 64 LKVEHVYDNGSVYSGWGLDIT 84
L VE VYDNGSV++ L++T
Sbjct 190 LVVESVYDNGSVFNPEVLNLT 210
> YLR340w
Length=312
Score = 86.7 bits (213), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 42/83 (50%), Positives = 56/83 (67%), Gaps = 0/83 (0%)
Query 4 STQTDPGSTGFFQALGISTKIVKGQIEIQQAVQLIRRGERVSASAATLLHKLSVKPFTYG 63
+T +PG T FFQALG+ TKI +G IEI V+++ G +V S A+LL+ L++ PFT+G
Sbjct 125 NTGMEPGKTSFFQALGVPTKIARGTIEIVSDVKVVDAGNKVGQSEASLLNLLNISPFTFG 184
Query 64 LKVEHVYDNGSVYSGWGLDITVE 86
L V VYDNG V+ LDIT E
Sbjct 185 LTVVQVYDNGQVFPSSILDITDE 207
> Hs20541776
Length=234
Score = 86.3 bits (212), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 42/84 (50%), Positives = 59/84 (70%), Gaps = 0/84 (0%)
Query 3 RSTQTDPGSTGFFQALGISTKIVKGQIEIQQAVQLIRRGERVSASAATLLHKLSVKPFTY 62
++T P FFQALGI+TKI +G EI VQLI+ G+RV AS ATLL+ ++ PF++
Sbjct 106 QNTGLGPEKISFFQALGITTKISRGTTEILSDVQLIKTGDRVGASEATLLNTPNISPFSF 165
Query 63 GLKVEHVYDNGSVYSGWGLDITVE 86
GL ++ V++NGS+Y+ LDIT E
Sbjct 166 GLVIQQVFNNGSIYNPEVLDITEE 189
> SPCC18.14c
Length=312
Score = 84.7 bits (208), Expect = 4e-17, Method: Composition-based stats.
Identities = 41/83 (49%), Positives = 56/83 (67%), Gaps = 0/83 (0%)
Query 4 STQTDPGSTGFFQALGISTKIVKGQIEIQQAVQLIRRGERVSASAATLLHKLSVKPFTYG 63
+T +PG T FFQALGI TKI +G IEI V L+ + +V S ATLL+ L++ PFTYG
Sbjct 125 NTGMEPGKTSFFQALGIPTKITRGTIEITSDVHLVSKDAKVGPSEATLLNMLNISPFTYG 184
Query 64 LKVEHVYDNGSVYSGWGLDITVE 86
+ V +YD G+V+S LD++ E
Sbjct 185 MDVLTIYDQGNVFSPEILDVSEE 207
> CE09655
Length=312
Score = 84.3 bits (207), Expect = 5e-17, Method: Composition-based stats.
Identities = 42/84 (50%), Positives = 56/84 (66%), Gaps = 0/84 (0%)
Query 3 RSTQTDPGSTGFFQALGISTKIVKGQIEIQQAVQLIRRGERVSASAATLLHKLSVKPFTY 62
++T P T FFQAL I TKI +G IEI V LI+ G++V AS + LL+ L V PF+Y
Sbjct 126 QNTGMGPEKTSFFQALQIPTKIARGTIEILNDVHLIKEGDKVGASESALLNMLGVTPFSY 185
Query 63 GLKVEHVYDNGSVYSGWGLDITVE 86
GL V VYD+G++Y+ LD+T E
Sbjct 186 GLVVRQVYDDGTLYTPEVLDMTTE 209
> 7296522
Length=317
Score = 83.6 bits (205), Expect = 8e-17, Method: Composition-based stats.
Identities = 43/84 (51%), Positives = 57/84 (67%), Gaps = 0/84 (0%)
Query 3 RSTQTDPGSTGFFQALGISTKIVKGQIEIQQAVQLIRRGERVSASAATLLHKLSVKPFTY 62
++T P T FFQAL I TKI KG IEI V +++ G++V AS ATLL+ L++ PF+Y
Sbjct 126 QNTGLGPEKTSFFQALSIPTKISKGTIEIINDVPILKPGDKVGASEATLLNMLNISPFSY 185
Query 63 GLKVEHVYDNGSVYSGWGLDITVE 86
GL V VYD+GS++S LDI E
Sbjct 186 GLIVNQVYDSGSIFSPEILDIKPE 209
> Hs22057309
Length=214
Score = 82.0 bits (201), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 41/84 (48%), Positives = 57/84 (67%), Gaps = 0/84 (0%)
Query 3 RSTQTDPGSTGFFQALGISTKIVKGQIEIQQAVQLIRRGERVSASAATLLHKLSVKPFTY 62
++T P T FFQALGI+TKI +G IEI VQLI+ G++V A+ ATLL+ S+ P ++
Sbjct 26 QNTGLGPEKTSFFQALGITTKISRGTIEILSDVQLIKTGDKVGANKATLLNTPSISPSSF 85
Query 63 GLKVEHVYDNGSVYSGWGLDITVE 86
L ++ DNGS+Y+ LDIT E
Sbjct 86 ALVIQQASDNGSIYNPEVLDITEE 109
> ECU07g0820
Length=290
Score = 72.0 bits (175), Expect = 3e-13, Method: Composition-based stats.
Identities = 33/82 (40%), Positives = 51/82 (62%), Gaps = 0/82 (0%)
Query 5 TQTDPGSTGFFQALGISTKIVKGQIEIQQAVQLIRRGERVSASAATLLHKLSVKPFTYGL 64
T P T +FQALGI+TKI KG++EI +++ G++V S A LL L++KPF Y +
Sbjct 150 TGMTPDKTSYFQALGIATKITKGKVEIISPYKVLSEGDKVGPSQANLLGMLNIKPFCYKM 209
Query 65 KVEHVYDNGSVYSGWGLDITVE 86
+ +Y++G +Y +DI E
Sbjct 210 TMHQIYEDGVIYDSSLIDIGEE 231
> Hs17437153_1
Length=334
Score = 70.5 bits (171), Expect = 7e-13, Method: Composition-based stats.
Identities = 37/69 (53%), Positives = 50/69 (72%), Gaps = 4/69 (5%)
Query 18 LGISTKIVKGQIEIQQAVQLIRRGERVSASAATLLHKLSVKPFTYGLKVEHVYDNGSVYS 77
LGI+TKI +G EI VQLI+ G++V AS ATL L++ PF +GL ++ V+DNGS+Y+
Sbjct 236 LGITTKISRGATEILSDVQLIKTGDKVGASEATL---LNISPF-FGLVIQQVFDNGSIYN 291
Query 78 GWGLDITVE 86
GLDIT E
Sbjct 292 PEGLDITEE 300
> Hs22061057
Length=184
Score = 62.4 bits (150), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 30/60 (50%), Positives = 42/60 (70%), Gaps = 0/60 (0%)
Query 27 GQIEIQQAVQLIRRGERVSASAATLLHKLSVKPFTYGLKVEHVYDNGSVYSGWGLDITVE 86
G EI VQLI+ G +V AS ATLL+ ++ PF++GL ++ V+DNGS+Y+ LDIT E
Sbjct 88 GTTEILSDVQLIKTGVKVGASEATLLNMRNISPFSFGLVIQQVFDNGSIYNTEVLDITEE 147
> Hs17437661
Length=122
Score = 61.2 bits (147), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 32/74 (43%), Positives = 51/74 (68%), Gaps = 1/74 (1%)
Query 3 RSTQTDPGSTGFFQALGISTKIVKGQIEIQQAVQLIRRGERVSASAATLLHKL-SVKPFT 61
++T P + FFQALGI+TKI + I I VQL+R G+++ AS ATLL+ L + + F+
Sbjct 46 QNTSLGPKKSFFFQALGITTKISRVTIGILSDVQLMRTGDKLEASEATLLNMLNTAQLFS 105
Query 62 YGLKVEHVYDNGSV 75
+GL ++ V++N S+
Sbjct 106 FGLIIKRVFNNSSI 119
> Hs22060831
Length=116
Score = 53.9 bits (128), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 27/72 (37%), Positives = 45/72 (62%), Gaps = 2/72 (2%)
Query 15 FQALGISTKIVKGQIEIQQAVQLIRRGERVSASAATLLHKLSVKPFTYGLKVEHVYDNGS 74
F A G TK+ +G +EI V LI+ G++ AS ATL++ ++ P ++ L ++ V+DN S
Sbjct 37 FSASG--TKMFRGSVEILSEVPLIKTGDKAGASEATLVNMWNISPSSFELIIQPVFDNCS 94
Query 75 VYSGWGLDITVE 86
+Y+ LD+ E
Sbjct 95 IYNPEVLDMAEE 106
> Hs22060836
Length=137
Score = 53.9 bits (128), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 27/72 (37%), Positives = 45/72 (62%), Gaps = 2/72 (2%)
Query 15 FQALGISTKIVKGQIEIQQAVQLIRRGERVSASAATLLHKLSVKPFTYGLKVEHVYDNGS 74
F A G TK+ +G +EI V LI+ G++ AS ATL++ ++ P ++ L ++ V+DN S
Sbjct 58 FSASG--TKMFRGSVEILSEVPLIKTGDKAGASEATLVNMWNISPSSFELIIQPVFDNCS 115
Query 75 VYSGWGLDITVE 86
+Y+ LD+ E
Sbjct 116 IYNPEVLDMAEE 127
> Hs17443776
Length=158
Score = 32.3 bits (72), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 17/42 (40%), Positives = 25/42 (59%), Gaps = 7/42 (16%)
Query 3 RSTQTDPGSTGFFQALGISTKIVKGQIEIQQAVQLIRRGERV 44
++T +P T FFQAL I+T EI VQLI+ G+++
Sbjct 75 QNTGLEPKKTSFFQALSITT-------EILSDVQLIKTGDKI 109
> YKL009w
Length=236
Score = 31.6 bits (70), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 26/55 (47%), Gaps = 0/55 (0%)
Query 20 ISTKIVKGQIEIQQAVQLIRRGERVSASAATLLHKLSVKPFTYGLKVEHVYDNGS 74
I TKI G+I I + GE++ A +L + + + +KV YDN S
Sbjct 171 IPTKIKAGKITIDSPYLVCTEGEKLDVRQALILKQFGIAASEFKVKVSAYYDNDS 225
> CE28326
Length=230
Score = 30.8 bits (68), Expect = 0.60, Method: Compositional matrix adjust.
Identities = 19/42 (45%), Positives = 25/42 (59%), Gaps = 4/42 (9%)
Query 10 GSTGFFQALGISTKIVKGQIEIQQAVQLIRRGERVSASAATL 51
GSTGFF ++ + G I QQA +IR ER A+AAT+
Sbjct 12 GSTGFF----VAHFMCAGFINQQQAEIIIRVPERPVAAAATM 49
> Hs22063216
Length=593
Score = 30.8 bits (68), Expect = 0.73, Method: Composition-based stats.
Identities = 21/63 (33%), Positives = 32/63 (50%), Gaps = 8/63 (12%)
Query 15 FQALGISTKIVK-------GQI-EIQQAVQLIRRGERVSASAATLLHKLSVKPFTYGLKV 66
FQ LG+ + V+ GQ E Q VQ + G R A +A + K +V+P+T ++
Sbjct 218 FQVLGLEVRSVRLGGCPDAGQTPETQPPVQSLFSGHRNLAPSARAMEKKNVQPWTLAERM 277
Query 67 EHV 69
E V
Sbjct 278 ETV 280
> CE04360
Length=220
Score = 29.3 bits (64), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 24/43 (55%), Gaps = 0/43 (0%)
Query 16 QALGISTKIVKGQIEIQQAVQLIRRGERVSASAATLLHKLSVK 58
+ LG+ TK+ KG I + Q ++ + GE ++ A +L VK
Sbjct 155 RKLGLPTKLDKGVITLYQQFEVCKEGEPLTVEQAKILKHFEVK 197
> CE10064
Length=501
Score = 28.5 bits (62), Expect = 2.9, Method: Composition-based stats.
Identities = 16/64 (25%), Positives = 31/64 (48%), Gaps = 6/64 (9%)
Query 10 GSTGFFQALGISTKIVKGQIEIQQAVQLIRRGERVSASAATLLHKLSVKPFTYGLKVEHV 69
G+ F+ +S + +++ + ++++R G R+ A L+ F Y L EH+
Sbjct 404 GTFALFETAHLSLHSEQATLDLVKCLEMVRNG-RIHAC-----QNLTQFSFVYTLLAEHI 457
Query 70 YDNG 73
DNG
Sbjct 458 LDNG 461
> SPAC18G6.07c
Length=359
Score = 26.9 bits (58), Expect = 8.5, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 29 IEIQQAVQLIRRGERVSASAATLLHKLSVKPFTYGLKVEHVYDN 72
IE+ +V++ R +R S LLHKLS++ K+ V N
Sbjct 229 IEVNPSVRIPRTFKRFSGLMVQLLHKLSIRSVNGNEKLLKVIKN 272
Lambda K H
0.317 0.133 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1190857334
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40