bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_8412_orf1
Length=88
Score E
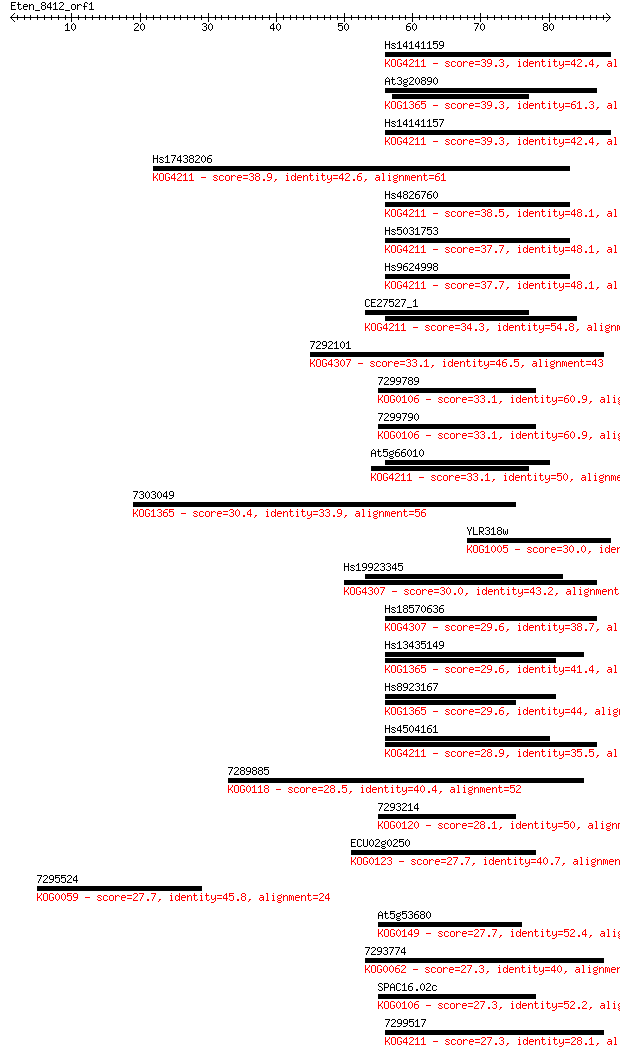
Sequences producing significant alignments: (Bits) Value
Hs14141159 39.3 0.002
At3g20890 39.3 0.002
Hs14141157 39.3 0.002
Hs17438206 38.9 0.002
Hs4826760 38.5 0.003
Hs5031753 37.7 0.005
Hs9624998 37.7 0.005
CE27527_1 34.3 0.052
7292101 33.1 0.12
7299789 33.1 0.13
7299790 33.1 0.14
At5g66010 33.1 0.14
7303049 30.4 0.85
YLR318w 30.0 1.0
Hs19923345 30.0 1.1
Hs18570636 29.6 1.4
Hs13435149 29.6 1.5
Hs8923167 29.6 1.5
Hs4504161 28.9 2.4
7289885 28.5 2.9
7293214 28.1 4.1
ECU02g0250 27.7 5.0
7295524 27.7 5.2
At5g53680 27.7 5.4
7293774 27.3 6.3
SPAC16.02c 27.3 6.9
7299517 27.3 7.5
> Hs14141159
Length=331
Score = 39.3 bits (90), Expect = 0.002, Method: Composition-based stats.
Identities = 14/33 (42%), Positives = 26/33 (78%), Gaps = 0/33 (0%)
Query 56 LRVRGLPFGARKEDLLEFFKGYKLAPSEDSIIM 88
+R+RGLPFG KE++++FF+G ++ P+ ++ M
Sbjct 18 VRLRGLPFGCSKEEIVQFFQGLEIVPNGITLTM 50
> At3g20890
Length=350
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/31 (61%), Positives = 25/31 (80%), Gaps = 2/31 (6%)
Query 56 LRVRGLPFGARKEDLLEFFKGYKLAPSEDSI 86
LR+RGLPF A KED+L+FFK ++L SED +
Sbjct 264 LRLRGLPFSAGKEDILDFFKDFEL--SEDFV 292
Score = 27.7 bits (60), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 11/20 (55%), Positives = 15/20 (75%), Gaps = 0/20 (0%)
Query 57 RVRGLPFGARKEDLLEFFKG 76
R+RGLPF + D++EFF G
Sbjct 118 RLRGLPFDCAELDVVEFFHG 137
> Hs14141157
Length=346
Score = 39.3 bits (90), Expect = 0.002, Method: Composition-based stats.
Identities = 14/33 (42%), Positives = 26/33 (78%), Gaps = 0/33 (0%)
Query 56 LRVRGLPFGARKEDLLEFFKGYKLAPSEDSIIM 88
+R+RGLPFG KE++++FF+G ++ P+ ++ M
Sbjct 18 VRLRGLPFGCSKEEIVQFFQGLEIVPNGITLTM 50
> Hs17438206
Length=126
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 26/79 (32%), Positives = 41/79 (51%), Gaps = 18/79 (22%)
Query 22 LNSSGEFKLA-----KTAAHS-AQAFRCMSILHD-------RNRPPR-----LRVRGLPF 63
L S E KLA +T HS +AF+ ++ D RN P +++RGLPF
Sbjct 36 LESENEVKLALKKDRETMGHSFVEAFKSNNVEMDWVLKHTGRNSPDTANDGFVQLRGLPF 95
Query 64 GARKEDLLEFFKGYKLAPS 82
KE++++FF G ++ P+
Sbjct 96 ECSKEEIVQFFSGLEIVPN 114
> Hs4826760
Length=415
Score = 38.5 bits (88), Expect = 0.003, Method: Composition-based stats.
Identities = 13/27 (48%), Positives = 22/27 (81%), Gaps = 0/27 (0%)
Query 56 LRVRGLPFGARKEDLLEFFKGYKLAPS 82
+R+RGLPFG KE++++FF G ++ P+
Sbjct 113 VRLRGLPFGCTKEEIVQFFSGLEIVPN 139
> Hs5031753
Length=449
Score = 37.7 bits (86), Expect = 0.005, Method: Composition-based stats.
Identities = 13/27 (48%), Positives = 22/27 (81%), Gaps = 0/27 (0%)
Query 56 LRVRGLPFGARKEDLLEFFKGYKLAPS 82
+R+RGLPFG KE++++FF G ++ P+
Sbjct 113 VRLRGLPFGCSKEEIVQFFSGLEIVPN 139
> Hs9624998
Length=449
Score = 37.7 bits (86), Expect = 0.005, Method: Composition-based stats.
Identities = 13/27 (48%), Positives = 22/27 (81%), Gaps = 0/27 (0%)
Query 56 LRVRGLPFGARKEDLLEFFKGYKLAPS 82
+R+RGLPFG KE++++FF G ++ P+
Sbjct 113 VRLRGLPFGCSKEEIVQFFSGLEIVPN 139
> CE27527_1
Length=607
Score = 34.3 bits (77), Expect = 0.052, Method: Composition-based stats.
Identities = 15/27 (55%), Positives = 20/27 (74%), Gaps = 3/27 (11%)
Query 53 PPR---LRVRGLPFGARKEDLLEFFKG 76
PPR +R+RGLPF A ++D+ EFF G
Sbjct 60 PPRSQYIRLRGLPFNATEKDIHEFFAG 86
Score = 30.8 bits (68), Expect = 0.60, Method: Composition-based stats.
Identities = 10/28 (35%), Positives = 22/28 (78%), Gaps = 0/28 (0%)
Query 56 LRVRGLPFGARKEDLLEFFKGYKLAPSE 83
+R+RG+P+ +++D+ +FF+G + P+E
Sbjct 159 IRLRGVPWSCKEDDVRKFFEGLEPPPAE 186
> 7292101
Length=985
Score = 33.1 bits (74), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 20/45 (44%), Positives = 29/45 (64%), Gaps = 4/45 (8%)
Query 45 SILHDR-NRPP-RLRVRGLPFGARKEDLLEFFKGYKLAPSEDSII 87
SI+ D+ NRP + +R +PF A +D++ FF YKL+P D II
Sbjct 898 SIIPDKFNRPGCVVAMRNVPFKAELKDIMRFFSDYKLSP--DDII 940
> 7299789
Length=329
Score = 33.1 bits (74), Expect = 0.13, Method: Composition-based stats.
Identities = 14/23 (60%), Positives = 17/23 (73%), Gaps = 0/23 (0%)
Query 55 RLRVRGLPFGARKEDLLEFFKGY 77
R+ V GLP+G R+ DL FFKGY
Sbjct 5 RVYVGGLPYGVRERDLERFFKGY 27
> 7299790
Length=132
Score = 33.1 bits (74), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 14/23 (60%), Positives = 17/23 (73%), Gaps = 0/23 (0%)
Query 55 RLRVRGLPFGARKEDLLEFFKGY 77
R+ V GLP+G R+ DL FFKGY
Sbjct 5 RVYVGGLPYGVRERDLERFFKGY 27
> At5g66010
Length=248
Score = 33.1 bits (74), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 12/24 (50%), Positives = 18/24 (75%), Gaps = 0/24 (0%)
Query 56 LRVRGLPFGARKEDLLEFFKGYKL 79
L++RGLP+ K ++EFF GYK+
Sbjct 161 LKMRGLPYSVNKPQIIEFFSGYKV 184
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 12/23 (52%), Positives = 15/23 (65%), Gaps = 0/23 (0%)
Query 54 PRLRVRGLPFGARKEDLLEFFKG 76
P +R+RGLPF D+ EFF G
Sbjct 42 PVVRLRGLPFNCADIDIFEFFAG 64
> 7303049
Length=557
Score = 30.4 bits (67), Expect = 0.85, Method: Composition-based stats.
Identities = 19/58 (32%), Positives = 31/58 (53%), Gaps = 8/58 (13%)
Query 19 LNVLNSSGE--FKLAKTAAHSAQAFRCMSILHDRNRPPRLRVRGLPFGARKEDLLEFF 74
+ V +SGE +A A++ AQAF + +R+RGLP+ A + +L+FF
Sbjct 15 IEVYRASGEDFLAIAGGASNEAQAFL------SKGAQVIIRMRGLPYDATAKQVLDFF 66
> YLR318w
Length=884
Score = 30.0 bits (66), Expect = 1.0, Method: Composition-based stats.
Identities = 10/21 (47%), Positives = 18/21 (85%), Gaps = 0/21 (0%)
Query 68 EDLLEFFKGYKLAPSEDSIIM 88
+DLLEF+ +K +PS+D++I+
Sbjct 646 DDLLEFYSEFKASPSQDTLIL 666
> Hs19923345
Length=932
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 10/29 (34%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 53 PPRLRVRGLPFGARKEDLLEFFKGYKLAP 81
P ++V+ +PF +++L+FF GY++ P
Sbjct 855 PTVIKVQNMPFTVSIDEILDFFYGYQVIP 883
Score = 28.1 bits (61), Expect = 4.2, Method: Composition-based stats.
Identities = 17/42 (40%), Positives = 25/42 (59%), Gaps = 7/42 (16%)
Query 50 RNRPPR-----LRVRGLPFGARKEDLLEFFKGYKLAPSEDSI 86
R+R P + ++GLPF A + +++FFK KL EDSI
Sbjct 421 RSRSPHEAGFCVYLKGLPFEAENKHVIDFFK--KLDIVEDSI 460
> Hs18570636
Length=1001
Score = 29.6 bits (65), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 12/31 (38%), Positives = 20/31 (64%), Gaps = 0/31 (0%)
Query 56 LRVRGLPFGARKEDLLEFFKGYKLAPSEDSI 86
+++ LPF A ++L+FF GY++ P SI
Sbjct 927 IKIMNLPFKANVNEILDFFHGYRIIPDSVSI 957
> Hs13435149
Length=727
Score = 29.6 bits (65), Expect = 1.5, Method: Composition-based stats.
Identities = 12/29 (41%), Positives = 22/29 (75%), Gaps = 1/29 (3%)
Query 56 LRVRGLPFGARKEDLLEFFKGYKLAPSED 84
+R++G+P+ A +DLL F+ Y+L P++D
Sbjct 676 VRMQGVPYTAGMKDLLSVFQAYQL-PADD 703
Score = 29.3 bits (64), Expect = 1.8, Method: Composition-based stats.
Identities = 11/25 (44%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 56 LRVRGLPFGARKEDLLEFFKGYKLA 80
+R RGLP+ + +D+ FFKG +A
Sbjct 259 VRARGLPWQSSDQDVARFFKGLNVA 283
> Hs8923167
Length=358
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 11/25 (44%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 56 LRVRGLPFGARKEDLLEFFKGYKLA 80
+R RGLP+ + +D+ FFKG +A
Sbjct 67 VRARGLPWQSSDQDIARFFKGLNIA 91
Score = 28.1 bits (61), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 10/19 (52%), Positives = 15/19 (78%), Gaps = 0/19 (0%)
Query 56 LRVRGLPFGARKEDLLEFF 74
+R+RGLP+ A ED+L+F
Sbjct 287 IRLRGLPYAATIEDILDFL 305
> Hs4504161
Length=424
Score = 28.9 bits (63), Expect = 2.4, Method: Composition-based stats.
Identities = 9/24 (37%), Positives = 18/24 (75%), Gaps = 0/24 (0%)
Query 56 LRVRGLPFGARKEDLLEFFKGYKL 79
+R+RGLP+ ++D+++FF G +
Sbjct 196 VRLRGLPYSCNEKDIVDFFAGLNI 219
Score = 27.3 bits (59), Expect = 7.8, Method: Composition-based stats.
Identities = 11/31 (35%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 56 LRVRGLPFGARKEDLLEFFKGYKLAPSEDSI 86
+R +GLP+ ED+L FF ++ E+ I
Sbjct 96 IRAQGLPWSCTMEDVLNFFSDCRIRNGENGI 126
> 7289885
Length=507
Score = 28.5 bits (62), Expect = 2.9, Method: Composition-based stats.
Identities = 21/56 (37%), Positives = 27/56 (48%), Gaps = 6/56 (10%)
Query 33 TAAHSAQAFRCMSILHDRNRPPRLRVRGLPFGARKEDLLEFFKGYKLA----PSED 84
TA + + F SI H P + LPF A ++DL EFF+G L P ED
Sbjct 256 TAPRANRIFDDNSIPH--KAPFIAYINNLPFDANEDDLYEFFEGINLISLRLPRED 309
> 7293214
Length=416
Score = 28.1 bits (61), Expect = 4.1, Method: Composition-based stats.
Identities = 10/20 (50%), Positives = 15/20 (75%), Gaps = 0/20 (0%)
Query 55 RLRVRGLPFGARKEDLLEFF 74
RL V +PFG +E+++EFF
Sbjct 94 RLYVGNIPFGVTEEEMMEFF 113
> ECU02g0250
Length=246
Score = 27.7 bits (60), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 11/27 (40%), Positives = 19/27 (70%), Gaps = 0/27 (0%)
Query 51 NRPPRLRVRGLPFGARKEDLLEFFKGY 77
+R R+ V G+P + KE++L+FF+ Y
Sbjct 78 DRKKRVAVSGIPSDSSKEEVLKFFQYY 104
> 7295524
Length=1713
Score = 27.7 bits (60), Expect = 5.2, Method: Composition-based stats.
Identities = 11/24 (45%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 5 RKGIKCLNSQNLIKLNVLNSSGEF 28
+KG++ N QNL LN+L +G F
Sbjct 53 QKGVRYYNEQNLTDLNLLQKNGGF 76
> At5g53680
Length=169
Score = 27.7 bits (60), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 11/21 (52%), Positives = 15/21 (71%), Gaps = 0/21 (0%)
Query 55 RLRVRGLPFGARKEDLLEFFK 75
++ V GLP+ RKE L+ FFK
Sbjct 14 KIYVGGLPWTTRKEGLINFFK 34
> 7293774
Length=708
Score = 27.3 bits (59), Expect = 6.3, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 18/35 (51%), Gaps = 0/35 (0%)
Query 53 PPRLRVRGLPFGARKEDLLEFFKGYKLAPSEDSII 87
PP L + + F ED L FKG L+ + DS I
Sbjct 488 PPVLAISEVTFRYNPEDPLPIFKGVNLSATSDSRI 522
> SPAC16.02c
Length=365
Score = 27.3 bits (59), Expect = 6.9, Method: Composition-based stats.
Identities = 12/23 (52%), Positives = 17/23 (73%), Gaps = 0/23 (0%)
Query 55 RLRVRGLPFGARKEDLLEFFKGY 77
RL V +P A +ED+++FFKGY
Sbjct 5 RLFVGRIPPQATREDMMDFFKGY 27
> 7299517
Length=586
Score = 27.3 bits (59), Expect = 7.5, Method: Composition-based stats.
Identities = 9/32 (28%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 56 LRVRGLPFGARKEDLLEFFKGYKLAPSEDSII 87
+++RGLP+ ++ + EFF G + + I+
Sbjct 148 VKLRGLPYAVTEQQIEEFFSGLDIKTDREGIL 179
Lambda K H
0.324 0.139 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1184307974
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40