bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_8524_orf1
Length=64
Score E
Sequences producing significant alignments: (Bits) Value
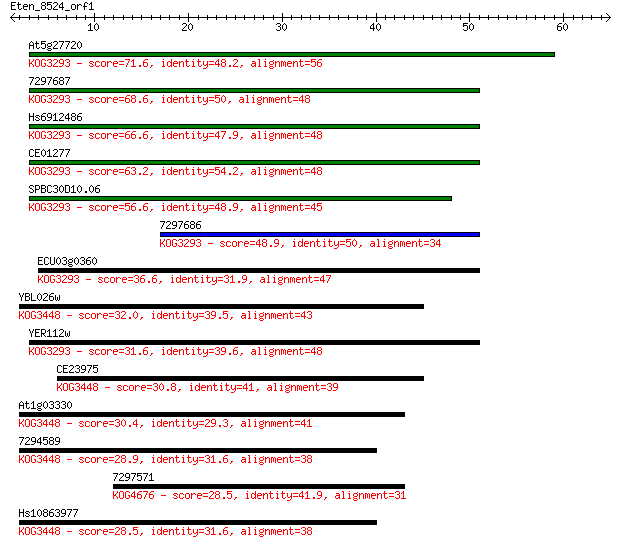
At5g27720 71.6 3e-13
7297687 68.6 3e-12
Hs6912486 66.6 1e-11
CE01277 63.2 1e-10
SPBC30D10.06 56.6 1e-08
7297686 48.9 2e-06
ECU03g0360 36.6 0.013
YBL026w 32.0 0.32
YER112w 31.6 0.41
CE23975 30.8 0.62
At1g03330 30.4 0.72
7294589 28.9 2.2
7297571 28.5 2.8
Hs10863977 28.5 3.0
> At5g27720
Length=129
Score = 71.6 bits (174), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 27/56 (48%), Positives = 43/56 (76%), Gaps = 0/56 (0%)
Query 3 FMNLHLKNSVCTSKDGERFFKVAECFIRGNTIKYARLQEELVALVKDENPKAGTSP 58
+MN+HL+ +CTSKDG+RF+++ EC+IRGNTIKY R+ +E++ V++E + P
Sbjct 35 WMNIHLREVICTSKDGDRFWRMPECYIRGNTIKYLRVPDEVIDKVQEEKTRTDRKP 90
> 7297687
Length=153
Score = 68.6 bits (166), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 24/48 (50%), Positives = 43/48 (89%), Gaps = 0/48 (0%)
Query 3 FMNLHLKNSVCTSKDGERFFKVAECFIRGNTIKYARLQEELVALVKDE 50
+MN++L++ +CTSKDG+RF+++ EC+IRG+TIKY R+ +E++ +VK++
Sbjct 34 WMNINLRDVICTSKDGDRFWRMPECYIRGSTIKYLRIPDEVIDMVKED 81
> Hs6912486
Length=139
Score = 66.6 bits (161), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 23/48 (47%), Positives = 42/48 (87%), Gaps = 0/48 (0%)
Query 3 FMNLHLKNSVCTSKDGERFFKVAECFIRGNTIKYARLQEELVALVKDE 50
+MN++L+ +CTS+DG++F+++ EC+IRG+TIKY R+ +E++ +VK+E
Sbjct 35 WMNINLREVICTSRDGDKFWRMPECYIRGSTIKYLRIPDEIIDMVKEE 82
> CE01277
Length=123
Score = 63.2 bits (152), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 26/48 (54%), Positives = 39/48 (81%), Gaps = 0/48 (0%)
Query 3 FMNLHLKNSVCTSKDGERFFKVAECFIRGNTIKYARLQEELVALVKDE 50
+MN+HL + + TSKDG++FFK++E ++RG+TIKY R+ E +V LVK E
Sbjct 36 WMNIHLVDVIFTSKDGDKFFKMSEAYVRGSTIKYLRIPETVVDLVKTE 83
> SPBC30D10.06
Length=121
Score = 56.6 bits (135), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 22/45 (48%), Positives = 35/45 (77%), Gaps = 0/45 (0%)
Query 3 FMNLHLKNSVCTSKDGERFFKVAECFIRGNTIKYARLQEELVALV 47
+MNL L+ + T DG++FF++ EC+IRGN IKY R+Q+E+++ V
Sbjct 35 YMNLTLREVIRTMPDGDKFFRLPECYIRGNNIKYLRIQDEVLSQV 79
> 7297686
Length=145
Score = 48.9 bits (115), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 17/34 (50%), Positives = 30/34 (88%), Gaps = 0/34 (0%)
Query 17 DGERFFKVAECFIRGNTIKYARLQEELVALVKDE 50
DG+RF+++ EC+IRG+TIKY R+ +E++ +VK++
Sbjct 40 DGDRFWRMPECYIRGSTIKYLRIPDEVIDMVKED 73
> ECU03g0360
Length=88
Score = 36.6 bits (83), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 15/47 (31%), Positives = 30/47 (63%), Gaps = 1/47 (2%)
Query 4 MNLHLKNSVCTSKDGERFFKVAECFIRGNTIKYARLQEELVALVKDE 50
MNLHLK+ +DG F + EC+++G +I+ +L+ +++ ++E
Sbjct 36 MNLHLKSVTIQKEDGSSIF-INECYLKGTSIRLVKLESKILLEQREE 81
> YBL026w
Length=95
Score = 32.0 bits (71), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 24/44 (54%), Gaps = 1/44 (2%)
Query 2 RFMNLHLKNSVCTS-KDGERFFKVAECFIRGNTIKYARLQEELV 44
+F+NL L N CT K V FIRG+T++Y L + +V
Sbjct 34 QFLNLKLDNISCTDEKKYPHLGSVRNIFIRGSTVRYVYLNKNMV 77
> YER112w
Length=187
Score = 31.6 bits (70), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 19/58 (32%), Positives = 30/58 (51%), Gaps = 10/58 (17%)
Query 3 FMNLHLKN-------SVCTSKD---GERFFKVAECFIRGNTIKYARLQEELVALVKDE 50
+MNL L N S S+D + K+ E +IRG IK+ +LQ+ ++ VK +
Sbjct 35 WMNLTLSNVTEYSEESAINSEDNAESSKAVKLNEIYIRGTFIKFIKLQDNIIDKVKQQ 92
> CE23975
Length=103
Score = 30.8 bits (68), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 24/42 (57%), Gaps = 3/42 (7%)
Query 6 LHLKNSVCTSKDGERF---FKVAECFIRGNTIKYARLQEELV 44
L++K + T D ERF V CFIRG+ ++Y +L + V
Sbjct 42 LNMKLTDITVSDPERFPHMVSVKNCFIRGSVVRYVQLPSDQV 83
> At1g03330
Length=93
Score = 30.4 bits (67), Expect = 0.72, Method: Compositional matrix adjust.
Identities = 12/42 (28%), Positives = 25/42 (59%), Gaps = 1/42 (2%)
Query 2 RFMNLHLKNSVCTSKDGE-RFFKVAECFIRGNTIKYARLQEE 42
+++N+ L+N+ +D V CFIRG+ ++Y +L ++
Sbjct 34 QYLNIKLENTRVVDQDKYPHMLSVRNCFIRGSVVRYVQLPKD 75
> 7294589
Length=95
Score = 28.9 bits (63), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 12/39 (30%), Positives = 21/39 (53%), Gaps = 1/39 (2%)
Query 2 RFMNLHLKNSVCTSKDGE-RFFKVAECFIRGNTIKYARL 39
+++N+ L + T D V CFIRG+ ++Y +L
Sbjct 34 QYLNIKLTDISVTDPDKYPHMLSVKNCFIRGSVVRYVQL 72
> 7297571
Length=513
Score = 28.5 bits (62), Expect = 2.8, Method: Composition-based stats.
Identities = 13/31 (41%), Positives = 17/31 (54%), Gaps = 0/31 (0%)
Query 12 VCTSKDGERFFKVAECFIRGNTIKYARLQEE 42
VC K+ R + ECF R +KYAR E+
Sbjct 168 VCDVKNEWRLDDLMECFQRAGEVKYARWAEK 198
> Hs10863977
Length=95
Score = 28.5 bits (62), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 12/39 (30%), Positives = 22/39 (56%), Gaps = 1/39 (2%)
Query 2 RFMNLHLKN-SVCTSKDGERFFKVAECFIRGNTIKYARL 39
+++N+ L + SV + V CFIRG+ ++Y +L
Sbjct 34 QYLNIKLTDISVTDPEKYPHMLSVKNCFIRGSVVRYVQL 72
Lambda K H
0.320 0.134 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1169706042
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40