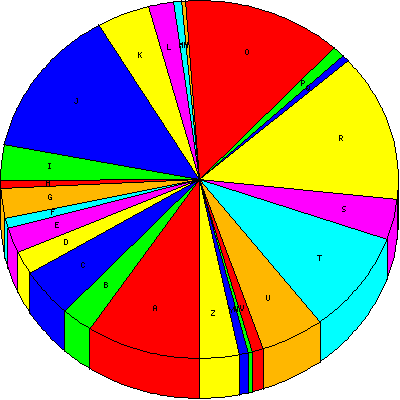
| A |
RNA processing and modification |
164 |
9.32 |
| B |
Chromatin structure and dynamics |
45 |
2.56 |
| C |
Energy production and conversion |
76 |
4.32 |
| D |
Cell cycle control, cell division, chromosome partitioning |
38 |
2.16 |
| E |
Amino acid transport and metabolism |
40 |
2.27 |
| F |
Nucleotide transport and metabolism |
15 |
0.85 |
| G |
Carbohydrate transport and metabolism |
48 |
2.73 |
| H |
Coenzyme transport and metabolism |
12 |
0.68 |
| I |
Lipid transport and metabolism |
57 |
3.24 |
| J |
Translation, ribosomal structure and biogenesis |
237 |
13.47 |
| K |
Transcription |
75 |
4.26 |
| L |
Replication, recombination and repair |
36 |
2.05 |
| M |
Cell wall/membrane/envelope biogenesis |
12 |
0.68 |
| N |
Cell motility |
5 |
0.28 |
| O |
Posttranslational modification, protein turnover, chaperones |
228 |
12.95 |
| P |
Inorganic ion transport and metabolism |
18 |
1.02 |
| Q |
Secondary metabolites biosynthesis, transport and catabolism |
11 |
0.62 |
| R |
General function prediction only |
234 |
13.30 |
| S |
Function unknown |
64 |
3.64 |
| T |
Signal transduction mechanisms |
163 |
9.26 |
| U |
Intracellular trafficking, secretion, and vesicular transport |
91 |
5.17 |
| V |
Defense mechanisms |
16 |
0.91 |
| W |
Extracellular structures |
6 |
0.34 |
| Y |
Nuclear structure |
13 |
0.74 |
| Z |
Cytoskeleton |
56 |
3.18 |
 Quantification of functional classes
Quantification of functional classes Quantification of functional classes
Quantification of functional classes