bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0009_orf1
Length=166
Score E
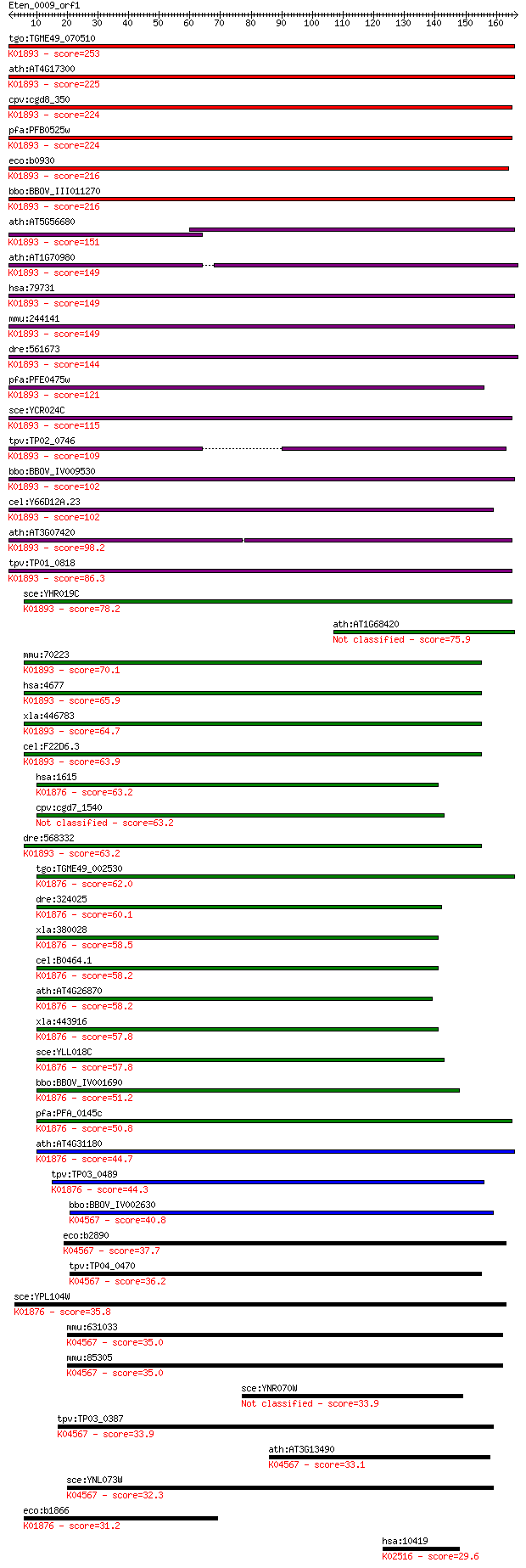
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_070510 asparaginyl-tRNA synthetase, putative (EC:6.... 253 3e-67
ath:AT4G17300 NS1; NS1; asparagine-tRNA ligase (EC:6.1.1.22); ... 225 5e-59
cpv:cgd8_350 asparaginyl-tRNA synthetase (NOB+tRNA synthase) ;... 224 1e-58
pfa:PFB0525w asparagine-tRNA ligase, putative; K01893 asparagi... 224 1e-58
eco:b0930 asnS, ECK0921, JW0913, lcs, tss; asparaginyl tRNA sy... 216 2e-56
bbo:BBOV_III011270 17.m07970; asparaginyl-tRNA synthetase fami... 216 4e-56
ath:AT5G56680 SYNC1; SYNC1; ATP binding / aminoacyl-tRNA ligas... 151 1e-36
ath:AT1G70980 SYNC3; SYNC3; ATP binding / aminoacyl-tRNA ligas... 149 3e-36
hsa:79731 NARS2, FLJ23441, SLM5; asparaginyl-tRNA synthetase 2... 149 4e-36
mmu:244141 Nars2, AI875199, MGC41336; asparaginyl-tRNA synthet... 149 5e-36
dre:561673 MGC153218; zgc:153218 (EC:6.1.1.22); K01893 asparag... 144 1e-34
pfa:PFE0475w asparagine-tRNA ligase, putative (EC:6.1.1.22); K... 121 1e-27
sce:YCR024C SLM5; Mitochondrial asparaginyl-tRNA synthetase (E... 115 5e-26
tpv:TP02_0746 asparaginyl-tRNA synthetase; K01893 asparaginyl-... 109 5e-24
bbo:BBOV_IV009530 23.m06478; asparaginyl-tRNA synthetase (EC:6... 102 5e-22
cel:Y66D12A.23 hypothetical protein; K01893 asparaginyl-tRNA s... 102 9e-22
ath:AT3G07420 NS2; NS2; asparagine-tRNA ligase (EC:6.1.1.22); ... 98.2 1e-20
tpv:TP01_0818 asparaginyl-tRNA synthetase; K01893 asparaginyl-... 86.3 4e-17
sce:YHR019C DED81; Ded81p (EC:6.1.1.22); K01893 asparaginyl-tR... 78.2 1e-14
ath:AT1G68420 asparaginyl-tRNA synthetase-related 75.9 6e-14
mmu:70223 Nars, 3010001M15Rik, AA960128, ASNRS, C78150; aspara... 70.1 3e-12
hsa:4677 NARS, ASNRS, NARS1; asparaginyl-tRNA synthetase (EC:6... 65.9 5e-11
xla:446783 nars, MGC80438; asparaginyl-tRNA synthetase (EC:6.1... 64.7 1e-10
cel:F22D6.3 nrs-1; asparagiNyl tRNA Synthetase family member (... 63.9 2e-10
hsa:1615 DARS, DKFZp781B11202, MGC111579; aspartyl-tRNA synthe... 63.2 3e-10
cpv:cgd7_1540 aspartate--tRNA ligase 63.2 4e-10
dre:568332 nars, fb57a10, si:dkey-274m14.2, wu:fb57a10; aspara... 63.2 4e-10
tgo:TGME49_002530 aspartyl-tRNA synthetase, putative (EC:6.1.1... 62.0 1e-09
dre:324025 dars, MGC154056, wu:fc17a11, zgc:154056; aspartyl-t... 60.1 3e-09
xla:380028 dars, MGC53970; aspartyl-tRNA synthetase (EC:6.1.1.... 58.5 1e-08
cel:B0464.1 drs-1; aspartyl(D) tRNA Synthetase family member (... 58.2 1e-08
ath:AT4G26870 aspartyl-tRNA synthetase, putative / aspartate--... 58.2 1e-08
xla:443916 dars, MGC80207; aspartyl-tRNA synthetase (EC:6.1.1.... 57.8 2e-08
sce:YLL018C DPS1; AspRS; aspartyl-tRNA synthetase (EC:6.1.1.12... 57.8 2e-08
bbo:BBOV_IV001690 21.m02784; aspartyl-tRNA synthetase (EC:6.1.... 51.2 1e-06
pfa:PFA_0145c aspartyl-tRNA synthetase, putative (EC:6.1.1.12)... 50.8 2e-06
ath:AT4G31180 aspartyl-tRNA synthetase, putative / aspartate--... 44.7 1e-04
tpv:TP03_0489 aspartyl-tRNA synthetase (EC:6.1.1.12); K01876 a... 44.3 2e-04
bbo:BBOV_IV002630 21.m02971; lysyl-tRNA synthetase (EC:6.1.1.6... 40.8 0.002
eco:b2890 lysS, asuD, ECK2885, herC, JW2858; lysine tRNA synth... 37.7 0.018
tpv:TP04_0470 lysyl-tRNA synthetase; K04567 lysyl-tRNA synthet... 36.2 0.045
sce:YPL104W MSD1, LPG5; Msd1p (EC:6.1.1.12); K01876 aspartyl-t... 35.8 0.074
mmu:631033 lysyl-tRNA synthetase-like; K04567 lysyl-tRNA synth... 35.0 0.10
mmu:85305 Kars, AA589550, AL024334, AL033315, AL033367, D8Ertd... 35.0 0.12
sce:YNR070W PDR18; Pdr18p 33.9 0.23
tpv:TP03_0387 lysyl-tRNA synthetase (EC:6.1.1.6); K04567 lysyl... 33.9 0.26
ath:AT3G13490 OVA5; OVA5 (OVULE ABORTION 5); ATP binding / ami... 33.1 0.38
sce:YNL073W MSK1; Mitochondrial lysine-tRNA synthetase, requir... 32.3 0.72
eco:b1866 aspS, ECK1867, JW1855, tls; aspartyl-tRNA synthetase... 31.2 1.5
hsa:10419 PRMT5, HRMT1L5, IBP72, JBP1, SKB1, SKB1Hs; protein a... 29.6 4.6
> tgo:TGME49_070510 asparaginyl-tRNA synthetase, putative (EC:6.1.1.22);
K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=676
Score = 253 bits (645), Expect = 3e-67, Method: Compositional matrix adjust.
Identities = 128/189 (67%), Positives = 147/189 (77%), Gaps = 24/189 (12%)
Query 1 LREIAHLRPRTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVS 60
LREIAHLRPR+ L+GAV R+RS L+MATHRFFQ+RGF YIHTPI+TASDCEGAGEMF VS
Sbjct 301 LREIAHLRPRSYLIGAVMRVRSNLAMATHRFFQDRGFLYIHTPIVTASDCEGAGEMFQVS 360
Query 61 TLLP-------EDPKKA----------------VPVNT-KGEIDYAKDFFSRHAYLTVSG 96
TLLP E+ KK +P+ K +DY++DFF R A+LTVSG
Sbjct 361 TLLPPPPPETRENEKKTDGAAPKDAAAAAAEPLIPLTKDKKGVDYSRDFFGRPAFLTVSG 420
Query 97 QLAVEPYCCALADVYTFGPTFRAENSHTSRHLAEFWMVEPEIAFADLQDDMQLAEQYVKF 156
QLAVEPYCC+L+DVYTFGPTFRAENSHTSRHLAEFWMVEPEIAFA L+D+M +AE YVKF
Sbjct 421 QLAVEPYCCSLSDVYTFGPTFRAENSHTSRHLAEFWMVEPEIAFATLEDNMVVAEAYVKF 480
Query 157 CVKSCLDNC 165
CV+ LDNC
Sbjct 481 CVQWVLDNC 489
> ath:AT4G17300 NS1; NS1; asparagine-tRNA ligase (EC:6.1.1.22);
K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=567
Score = 225 bits (573), Expect = 5e-59, Method: Compositional matrix adjust.
Identities = 106/171 (61%), Positives = 129/171 (75%), Gaps = 6/171 (3%)
Query 1 LREIAHLRPRTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVS 60
LR AHLRPRT GAVAR+R+ L+ ATH+FFQE GF ++ +PIITASDCEGAGE F V+
Sbjct 211 LRTKAHLRPRTNTFGAVARVRNTLAYATHKFFQESGFVWVASPIITASDCEGAGEQFCVT 270
Query 61 TLLPEDPKK------AVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCALADVYTFG 114
TL+P + A+P G ID+++DFF + A+LTVSGQL E Y AL+DVYTFG
Sbjct 271 TLIPSSHENTDTSIDAIPKTKGGLIDWSQDFFGKPAFLTVSGQLNGETYATALSDVYTFG 330
Query 115 PTFRAENSHTSRHLAEFWMVEPEIAFADLQDDMQLAEQYVKFCVKSCLDNC 165
PTFRAENS+TSRHLAEFWM+EPE+AFADL DDM A Y+++ VK LDNC
Sbjct 331 PTFRAENSNTSRHLAEFWMIEPELAFADLDDDMACATAYLQYVVKYVLDNC 381
> cpv:cgd8_350 asparaginyl-tRNA synthetase (NOB+tRNA synthase)
; K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=499
Score = 224 bits (570), Expect = 1e-58, Method: Compositional matrix adjust.
Identities = 101/172 (58%), Positives = 131/172 (76%), Gaps = 8/172 (4%)
Query 1 LREIAHLRPRTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVS 60
LRE+AHLRPR+Q +V RIR+ L++A H +FQ+ GF YIHTPIITA+DCEGAGEMF V+
Sbjct 142 LREVAHLRPRSQFFSSVMRIRNSLAIAIHEYFQKNGFMYIHTPIITAADCEGAGEMFQVT 201
Query 61 TLLPED--------PKKAVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCALADVYT 112
T+LP + P + + +DY KDFF + +YLTVSGQLA+E + C+++DVYT
Sbjct 202 TVLPPESKNNISNIPASKIEGSQDMAVDYKKDFFGKASYLTVSGQLALENFACSMSDVYT 261
Query 113 FGPTFRAENSHTSRHLAEFWMVEPEIAFADLQDDMQLAEQYVKFCVKSCLDN 164
FGPTFRAENSHT+RHLAEFWM+EPE+AFADL D+M+L E +K+ V+ L N
Sbjct 262 FGPTFRAENSHTTRHLAEFWMIEPEMAFADLSDNMKLGEGLLKYTVEYVLIN 313
> pfa:PFB0525w asparagine-tRNA ligase, putative; K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=610
Score = 224 bits (570), Expect = 1e-58, Method: Composition-based stats.
Identities = 110/219 (50%), Positives = 138/219 (63%), Gaps = 55/219 (25%)
Query 1 LREIAHLRPRTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVS 60
LRE+AHLRPR+ + +V RIR+ LS+ATH FFQ RGF YIHTP+IT SDCEG GEMF+V+
Sbjct 206 LREVAHLRPRSYFISSVIRIRNSLSIATHLFFQSRGFLYIHTPLITTSDCEGGGEMFTVT 265
Query 61 TLLPEDPK-KAVP-VNTKGE---------------------------------------- 78
TLL E+ +++P +N K +
Sbjct 266 TLLNENGDIRSIPRINLKNKKKEKREDILNEKNGKKDHMNDSLNNNTCNNNNNNGNSSSS 325
Query 79 -------------IDYAKDFFSRHAYLTVSGQLAVEPYCCALADVYTFGPTFRAENSHTS 125
IDY KDFFS+ A+LTVSGQL++E C ++ DVYTFGPTFRAENSHTS
Sbjct 326 NIVSSPQYEDNYLIDYKKDFFSKQAFLTVSGQLSLENLCSSMGDVYTFGPTFRAENSHTS 385
Query 126 RHLAEFWMVEPEIAFADLQDDMQLAEQYVKFCVKSCLDN 164
RHLAEFWM+EPEIAFADL D+M+LAE Y+K+C+ L+N
Sbjct 386 RHLAEFWMIEPEIAFADLYDNMELAEAYIKYCIDYVLNN 424
> eco:b0930 asnS, ECK0921, JW0913, lcs, tss; asparaginyl tRNA
synthetase (EC:6.1.1.22); K01893 asparaginyl-tRNA synthetase
[EC:6.1.1.22]
Length=466
Score = 216 bits (551), Expect = 2e-56, Method: Compositional matrix adjust.
Identities = 101/163 (61%), Positives = 128/163 (78%), Gaps = 4/163 (2%)
Query 1 LREIAHLRPRTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVS 60
LRE+AHLRPRT L+GAVAR+R L+ A HRFF E+GF ++ TP+ITASD EGAGEMF VS
Sbjct 121 LREVAHLRPRTNLIGAVARVRHTLAQALHRFFNEQGFFWVSTPLITASDTEGAGEMFRVS 180
Query 61 TLLPEDPKKAVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCALADVYTFGPTFRAE 120
TL E+ +P N +G++D+ KDFF + ++LTVSGQL E Y CAL+ +YTFGPTFRAE
Sbjct 181 TLDLEN----LPRNDQGKVDFDKDFFGKESFLTVSGQLNGETYACALSKIYTFGPTFRAE 236
Query 121 NSHTSRHLAEFWMVEPEIAFADLQDDMQLAEQYVKFCVKSCLD 163
NS+TSRHLAEFWM+EPE+AFA+L D LAE +K+ K+ L+
Sbjct 237 NSNTSRHLAEFWMLEPEVAFANLNDIAGLAEAMLKYVFKAVLE 279
> bbo:BBOV_III011270 17.m07970; asparaginyl-tRNA synthetase family
protein; K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=605
Score = 216 bits (549), Expect = 4e-56, Method: Compositional matrix adjust.
Identities = 103/180 (57%), Positives = 130/180 (72%), Gaps = 16/180 (8%)
Query 1 LREIAHLRPRTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVS 60
LRE+AHLRPR+ + +V R+RS L+MATH +FQ G Y+HTP+IT +DCEGAGE+F V+
Sbjct 217 LREVAHLRPRSYFISSVMRVRSALAMATHIYFQRLGCIYLHTPLITTTDCEGAGELFQVT 276
Query 61 TLLPE---------------DPKKAVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCC 105
T+L + DPKKA + + E+DY KDFF R AYLT SGQL+ E Y C
Sbjct 277 TMLEDCKTLGDLAKKMTAHGDPKKA-KILEEMEMDYKKDFFKRKAYLTCSGQLSAENYAC 335
Query 106 ALADVYTFGPTFRAENSHTSRHLAEFWMVEPEIAFADLQDDMQLAEQYVKFCVKSCLDNC 165
++ VYTFGPTFRAE+SHTSRHLAEFWMVEPEI F DL +M +AE+YV+F +K LD+C
Sbjct 336 SMGKVYTFGPTFRAEHSHTSRHLAEFWMVEPEIDFCDLAKNMDIAEEYVQFAIKYALDHC 395
> ath:AT5G56680 SYNC1; SYNC1; ATP binding / aminoacyl-tRNA ligase/
asparagine-tRNA ligase/ aspartate-tRNA ligase/ nucleic
acid binding / nucleotide binding (EC:6.1.1.22); K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=572
Score = 151 bits (381), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 73/106 (68%), Positives = 80/106 (75%), Gaps = 6/106 (5%)
Query 60 STLLPEDPKKAVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCALADVYTFGPTFRA 119
S L P PKK G IDY+KDFF R A+LTVSGQL VE Y CAL++VYTFGPTFRA
Sbjct 285 SRLRPGLPKK------DGNIDYSKDFFGRQAFLTVSGQLQVETYACALSNVYTFGPTFRA 338
Query 120 ENSHTSRHLAEFWMVEPEIAFADLQDDMQLAEQYVKFCVKSCLDNC 165
ENSHTSRHLAEFWMVEPEIAFADL+DDM AE YVK+ L+ C
Sbjct 339 ENSHTSRHLAEFWMVEPEIAFADLEDDMNCAEAYVKYMCNWLLEKC 384
Score = 92.8 bits (229), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 42/63 (66%), Positives = 49/63 (77%), Gaps = 0/63 (0%)
Query 1 LREIAHLRPRTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVS 60
LR++ HLR RT + AVARIR+ L+ ATH FFQE F YIHTPIIT SDCEGAGEMF +
Sbjct 154 LRDVLHLRSRTNSISAVARIRNALAFATHSFFQEHSFLYIHTPIITTSDCEGAGEMFQAT 213
Query 61 TLL 63
TL+
Sbjct 214 TLI 216
> ath:AT1G70980 SYNC3; SYNC3; ATP binding / aminoacyl-tRNA ligase/
asparagine-tRNA ligase/ aspartate-tRNA ligase/ nucleic
acid binding / nucleotide binding (EC:6.1.1.22); K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=571
Score = 149 bits (377), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 69/99 (69%), Positives = 77/99 (77%), Gaps = 1/99 (1%)
Query 68 KKAVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCALADVYTFGPTFRAENSHTSRH 127
K +P N G+IDY+ DFF R A+LTVSGQL VE Y CAL+ VYTFGPTFRAENSHTSRH
Sbjct 285 KPGLPKN-DGKIDYSNDFFGRQAFLTVSGQLQVETYACALSSVYTFGPTFRAENSHTSRH 343
Query 128 LAEFWMVEPEIAFADLQDDMQLAEQYVKFCVKSCLDNCS 166
LAEFWMVEPEIAFAD+ DDM AE YVK+ K +D C
Sbjct 344 LAEFWMVEPEIAFADIHDDMNCAEAYVKYMCKWLMDKCG 382
Score = 97.4 bits (241), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 44/63 (69%), Positives = 51/63 (80%), Gaps = 0/63 (0%)
Query 1 LREIAHLRPRTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVS 60
LR++ HLR RT L+ AVARIR+ L+ ATH FFQE F YIHTPIIT SDCEGAGEMF V+
Sbjct 151 LRDVLHLRSRTNLISAVARIRNALAFATHSFFQEHSFLYIHTPIITTSDCEGAGEMFQVT 210
Query 61 TLL 63
TL+
Sbjct 211 TLI 213
> hsa:79731 NARS2, FLJ23441, SLM5; asparaginyl-tRNA synthetase
2, mitochondrial (putative) (EC:6.1.1.22); K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=477
Score = 149 bits (376), Expect = 4e-36, Method: Compositional matrix adjust.
Identities = 80/166 (48%), Positives = 105/166 (63%), Gaps = 14/166 (8%)
Query 1 LREIAHLRPRTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVS 60
LR+ H R RT ++G++ RIRS+ + A H FF++ GF +IHTPIIT++D EGAGE+F
Sbjct 139 LRQYPHFRCRTNVLGSILRIRSEATAAIHSFFKDSGFVHIHTPIITSNDSEGAGELFQ-- 196
Query 61 TLLPEDPKKAVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCALADVYTFGPTFRAE 120
L K VP ++FF+ A+LTVSGQL +E A V+TFGPTFRAE
Sbjct 197 --LEPSGKLKVP---------EENFFNVPAFLTVSGQLHLEVMSGAFTQVFTFGPTFRAE 245
Query 121 NSHTSRHLAEFWMVEPEIAFAD-LQDDMQLAEQYVKFCVKSCLDNC 165
NS + RHLAEF+M+E EI+F D LQD MQ+ E+ K L C
Sbjct 246 NSQSRRHLAEFYMIEAEISFVDSLQDLMQVIEELFKATTMMVLSKC 291
> mmu:244141 Nars2, AI875199, MGC41336; asparaginyl-tRNA synthetase
2 (mitochondrial)(putative) (EC:6.1.1.22); K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=477
Score = 149 bits (375), Expect = 5e-36, Method: Compositional matrix adjust.
Identities = 78/166 (46%), Positives = 108/166 (65%), Gaps = 14/166 (8%)
Query 1 LREIAHLRPRTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVS 60
LR+ HLR RT +G++ R+RS+ + A H +F++ GF +IHTP++T++DCEGAGE+F V
Sbjct 139 LRQYPHLRCRTNALGSILRVRSEATAAIHSYFKDNGFVHIHTPVLTSNDCEGAGELFQV- 197
Query 61 TLLPEDPKKAVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCALADVYTFGPTFRAE 120
+P + KG + FF A+LTVSGQL +E A V+TFGPTFRAE
Sbjct 198 -----EPSSKI----KGP---KESFFDVPAFLTVSGQLHLEVMSGAFTQVFTFGPTFRAE 245
Query 121 NSHTSRHLAEFWMVEPEIAFAD-LQDDMQLAEQYVKFCVKSCLDNC 165
NS + RHLAEF+MVE EI+F + LQD MQ+ E+ K + L +C
Sbjct 246 NSQSRRHLAEFYMVEAEISFVESLQDLMQVMEELFKATTEMVLSHC 291
> dre:561673 MGC153218; zgc:153218 (EC:6.1.1.22); K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=485
Score = 144 bits (364), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 79/167 (47%), Positives = 102/167 (61%), Gaps = 13/167 (7%)
Query 1 LREIAHLRPRTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVS 60
+R+ HLR RT ++ RIRS+ + A FF++ G+ IHTP+IT++DCEGAGE+F V
Sbjct 146 IRQFPHLRCRTNAFSSMLRIRSEATAAIQSFFRDHGYVQIHTPVITSNDCEGAGELFQVE 205
Query 61 TLLPEDPKKAVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCALADVYTFGPTFRAE 120
P K + D FFS AYLTVSGQL +E A + VYTFGPTFRAE
Sbjct 206 -----------PTGDKKDSD-DPHFFSVPAYLTVSGQLHLEVMAGAFSAVYTFGPTFRAE 253
Query 121 NSHTSRHLAEFWMVEPEIAFADLQDD-MQLAEQYVKFCVKSCLDNCS 166
NS + RHLAEF+MVE EIAF + DD M++ E K + NC+
Sbjct 254 NSQSRRHLAEFYMVEAEIAFTESLDDLMKVMEDLFKAATEHVFSNCT 300
> pfa:PFE0475w asparagine-tRNA ligase, putative (EC:6.1.1.22);
K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=722
Score = 121 bits (303), Expect = 1e-27, Method: Composition-based stats.
Identities = 70/221 (31%), Positives = 110/221 (49%), Gaps = 71/221 (32%)
Query 1 LREIAHLRPRTQLMGAVARIRSQLSMATHRFFQER-GFQYIHTPIITASDCEGAGEMFSV 59
LR HLR RT+L ++ R++S + T F +++ + YI+TPI+T++DCEGAG++F
Sbjct 315 LRNYPHLRARTKLYSSLFRLKSDIIFETFNFLKKKKNYTYINTPILTSNDCEGAGKLFYA 374
Query 60 STLLPEDPKKAVPVNTKGEIDYAK----------------------DFFSRH-------- 89
+TL + K N + E D+ K ++ SRH
Sbjct 375 TTLFDQQIK-----NEQREEDHMKKGITNEIAGNIKEKCYYQNEHLEYISRHKSISKMEG 429
Query 90 -----------------------------------AYLTVSGQLAVEPYCCALADVYTFG 114
YL VS QL++E CC++ DV+T
Sbjct 430 FDEMLLKNVSSNDCISPNNTESYNTRFENDFFKKACYLNVSSQLSLECLCCSIGDVFTLN 489
Query 115 PTFRAENSHTSRHLAEFWMVEPEIAFADLQDDMQLAEQYVK 155
P+FRAENS+T RHL+EF M+E E+AF++L+D + ++E+Y+K
Sbjct 490 PSFRAENSNTFRHLSEFLMLEVELAFSNLKDIIDISEEYIK 530
> sce:YCR024C SLM5; Mitochondrial asparaginyl-tRNA synthetase
(EC:6.1.1.22); K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=492
Score = 115 bits (289), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 63/165 (38%), Positives = 93/165 (56%), Gaps = 16/165 (9%)
Query 1 LREIAHLRPRTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVS 60
LR + L+ RT + A+ R+RS + +FQ+ F + PI+T++DCEGAGE+F VS
Sbjct 137 LRSLPTLKYRTAYLSAILRLRSFVEFQFMLYFQKNHFTKVSPPILTSNDCEGAGELFQVS 196
Query 61 TLLPEDPKKAVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCALADVYTFGPTFRAE 120
T NT A +F + YLTVS QL +E +L+ +T P FRAE
Sbjct 197 T------------NTSPT---ASSYFGKPTYLTVSTQLHLEILALSLSRCWTLSPCFRAE 241
Query 121 NSHTSRHLAEFWMVEPEIAFADLQDDM-QLAEQYVKFCVKSCLDN 164
S T RHL+EFWM+E E+ F + +++ E +K +K+C+DN
Sbjct 242 KSDTPRHLSEFWMLEVEMCFVNSVNELTSFVETTIKHIIKACIDN 286
> tpv:TP02_0746 asparaginyl-tRNA synthetase; K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=710
Score = 109 bits (272), Expect = 5e-24, Method: Composition-based stats.
Identities = 44/73 (60%), Positives = 57/73 (78%), Gaps = 0/73 (0%)
Query 90 AYLTVSGQLAVEPYCCALADVYTFGPTFRAENSHTSRHLAEFWMVEPEIAFADLQDDMQL 149
+YLT SGQL+ E YCC++ VYTFGPTFRAENSHT+RHL+EFWM+EPE+ DL M++
Sbjct 444 SYLTCSGQLSAENYCCSMGSVYTFGPTFRAENSHTNRHLSEFWMIEPEMTLVDLPGLMEI 503
Query 150 AEQYVKFCVKSCL 162
E+++KF V L
Sbjct 504 TEEFIKFLVNYIL 516
Score = 92.8 bits (229), Expect = 5e-19, Method: Composition-based stats.
Identities = 39/63 (61%), Positives = 50/63 (79%), Gaps = 0/63 (0%)
Query 1 LREIAHLRPRTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVS 60
LRE AHLRPRT L+ AV R+RS LS+A H FFQ + F Y+++P+IT +DCEGAGE+F V+
Sbjct 277 LRENAHLRPRTYLISAVMRVRSSLSIAIHLFFQSKNFHYLNSPVITTADCEGAGELFQVT 336
Query 61 TLL 63
TL
Sbjct 337 TLF 339
> bbo:BBOV_IV009530 23.m06478; asparaginyl-tRNA synthetase (EC:6.1.1.22);
K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=557
Score = 102 bits (255), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 58/177 (32%), Positives = 85/177 (48%), Gaps = 40/177 (22%)
Query 1 LREIAHLRPRTQLMGAVARIRSQLSMATHRF------------FQERGFQYIHTPIITAS 48
LRE + R R +M A+ RIRS ++ + Q+R F + TP++T
Sbjct 198 LREHVNFRSRNAVMQAIFRIRSAVTEGLYHIMKVCHVTGSLYSLQKRSFINVTTPLLTTV 257
Query 49 DCEGAGEMFSVSTLLPEDPKKAVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCALA 108
+CEG G++F EID +L+VSGQL +E C +
Sbjct 258 NCEGMGDIF--------------------EID--------RTFLSVSGQLELEALCSGFS 289
Query 109 DVYTFGPTFRAENSHTSRHLAEFWMVEPEIAFADLQDDMQLAEQYVKFCVKSCLDNC 165
V+ GP FRA+ S T RHL EFWM+E E+ DL+D + + Q + ++ LDNC
Sbjct 290 RVWKLGPAFRADRSDTPRHLCEFWMLEAEMVGPDLKDLINVIHQIIIGSARTILDNC 346
> cel:Y66D12A.23 hypothetical protein; K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=448
Score = 102 bits (253), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 61/163 (37%), Positives = 83/163 (50%), Gaps = 22/163 (13%)
Query 1 LREIAHLRPRTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSV- 59
LR+ HLR R A+ R RS + TH FF R F +I TP +T +DCEG GE+F V
Sbjct 102 LRKKTHLRARNSKFAALLRARSAIFRETHEFFMSRDFIHIDTPKLTRNDCEGGGEVFDVV 161
Query 60 ---STLLPEDPKKAVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCALADVYTFGPT 116
ST + K P+ +L+VS QL +E L VY GP+
Sbjct 162 TTTSTEAGDGGLKEEPM-----------------FLSVSSQLHLEAMVSRLQRVYILGPS 204
Query 117 FRAENSHTSRHLAEFWMVEPEIAFADLQDDM-QLAEQYVKFCV 158
FRAE + HL+EF M+E E+AF + D+M L E Y++ +
Sbjct 205 FRAERQQSHAHLSEFHMLEAEVAFVETIDEMCTLVESYIQHMI 247
> ath:AT3G07420 NS2; NS2; asparagine-tRNA ligase (EC:6.1.1.22);
K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=638
Score = 98.2 bits (243), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 45/87 (51%), Positives = 58/87 (66%), Gaps = 0/87 (0%)
Query 78 EIDYAKDFFSRHAYLTVSGQLAVEPYCCALADVYTFGPTFRAENSHTSRHLAEFWMVEPE 137
++D++KDFF R YLT SG+ +E Y AL VYTFGP F A+ +RHLAE W VE E
Sbjct 365 KLDFSKDFFGRDTYLTASGRFHLESYASALGKVYTFGPRFIADKIDNARHLAEKWNVETE 424
Query 138 IAFADLQDDMQLAEQYVKFCVKSCLDN 164
+AFA+L D M A++Y KF K L+N
Sbjct 425 MAFAELDDAMDCADEYFKFLCKYVLEN 451
Score = 72.8 bits (177), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 36/79 (45%), Positives = 50/79 (63%), Gaps = 5/79 (6%)
Query 1 LREIAHLRPRTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVS 60
LR+ +H RPRT +G+V R+ S L++A+H F Q GFQY+ P+IT + G GEMF V+
Sbjct 224 LRDFSHFRPRTTTVGSVTRVHSALTLASHTFLQYHGFQYVQVPVITTT--TGFGEMFRVT 281
Query 61 TLL---PEDPKKAVPVNTK 76
TLL + +K PV K
Sbjct 282 TLLGKTDDKEEKKPPVQEK 300
> tpv:TP01_0818 asparaginyl-tRNA synthetase; K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=558
Score = 86.3 bits (212), Expect = 4e-17, Method: Composition-based stats.
Identities = 50/164 (30%), Positives = 72/164 (43%), Gaps = 29/164 (17%)
Query 1 LREIAHLRPRTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVS 60
LR+ H R R + ++ IRS + H+FF F + TP +T +CE
Sbjct 230 LRKYTHFRFRNKQFRSIIAIRSAIKSYIHQFFTSNKFTEVDTPCLTLMNCENLA------ 283
Query 61 TLLPEDPKKAVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCALADVYTFGPTFRAE 120
P +PVN L+ +GQ+ +E C L V+ GP+FR E
Sbjct 284 ------PLTNIPVN-----------------LSSTGQMELEFACYGLGKVWKVGPSFRGE 320
Query 121 NSHTSRHLAEFWMVEPEIAFADLQDDMQLAEQYVKFCVKSCLDN 164
S T RH EFWM+E EI A L + L ++ ++ C L N
Sbjct 321 RSDTIRHANEFWMIEAEIPDATLTGMINLIQELIRGCATYILQN 364
> sce:YHR019C DED81; Ded81p (EC:6.1.1.22); K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=554
Score = 78.2 bits (191), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 46/159 (28%), Positives = 72/159 (45%), Gaps = 24/159 (15%)
Query 6 HLRPRTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTLLPE 65
HL R + AV ++R+ L + R + E + P + + EG +F +
Sbjct 239 HLALRGDALSAVMKVRAALLKSVRRVYDEEHLTEVTPPCMVQTQVEGGSTLFKM------ 292
Query 66 DPKKAVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCALADVYTFGPTFRAENSHTS 125
+++ AYLT S QL +E +L DVYT +FRAE SHT
Sbjct 293 ------------------NYYGEEAYLTQSSQLYLETCLASLGDVYTIQESFRAEKSHTR 334
Query 126 RHLAEFWMVEPEIAFADLQDDMQLAEQYVKFCVKSCLDN 164
RHL+E+ +E E+AF D +Q E + V+ L++
Sbjct 335 RHLSEYTHIEAELAFLTFDDLLQHIETLIVKSVQYVLED 373
> ath:AT1G68420 asparaginyl-tRNA synthetase-related
Length=87
Score = 75.9 bits (185), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 35/59 (59%), Positives = 43/59 (72%), Gaps = 0/59 (0%)
Query 107 LADVYTFGPTFRAENSHTSRHLAEFWMVEPEIAFADLQDDMQLAEQYVKFCVKSCLDNC 165
+ DVYTF PTFRAE SHTSRHLAEFWMVE E+AFA +++ M +E VK + L+ C
Sbjct 1 MGDVYTFAPTFRAEKSHTSRHLAEFWMVEVELAFAGVEEAMNCSEAVVKDMCTTLLEKC 59
> mmu:70223 Nars, 3010001M15Rik, AA960128, ASNRS, C78150; asparaginyl-tRNA
synthetase (EC:6.1.1.22); K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=559
Score = 70.1 bits (170), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 44/149 (29%), Positives = 65/149 (43%), Gaps = 24/149 (16%)
Query 6 HLRPRTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTLLPE 65
H+ R + M + + RS ++ F +RG+ + TP + + EG +F +
Sbjct 245 HMMIRGENMSKILKARSMITRCFRDHFFDRGYCEVTTPTLVQTQVEGGATLFKL------ 298
Query 66 DPKKAVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCALADVYTFGPTFRAENSHTS 125
D+F A+LT S QL +E AL DV+ ++RAE S T
Sbjct 299 ------------------DYFGEEAFLTQSSQLYLETCLPALGDVFCIAQSYRAEQSRTR 340
Query 126 RHLAEFWMVEPEIAFADLQDDMQLAEQYV 154
RHLAEF VE E F +D + E V
Sbjct 341 RHLAEFTHVEAECPFLTFEDLLNRLEDLV 369
> hsa:4677 NARS, ASNRS, NARS1; asparaginyl-tRNA synthetase (EC:6.1.1.22);
K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=548
Score = 65.9 bits (159), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 42/149 (28%), Positives = 63/149 (42%), Gaps = 24/149 (16%)
Query 6 HLRPRTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTLLPE 65
H+ R + M + + RS ++ F +RG+ + P + + EG +F +
Sbjct 234 HMMIRGENMSKILKARSMVTRCFRDHFFDRGYYEVTPPTLVQTQVEGGATLFKL------ 287
Query 66 DPKKAVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCALADVYTFGPTFRAENSHTS 125
D+F A+LT S QL +E AL DV+ ++RAE S T
Sbjct 288 ------------------DYFGEEAFLTQSSQLYLETCLPALGDVFCIAQSYRAEQSRTR 329
Query 126 RHLAEFWMVEPEIAFADLQDDMQLAEQYV 154
RHLAE+ VE E F D + E V
Sbjct 330 RHLAEYTHVEAECPFLTFDDLLNRLEDLV 358
> xla:446783 nars, MGC80438; asparaginyl-tRNA synthetase (EC:6.1.1.22);
K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=559
Score = 64.7 bits (156), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 40/149 (26%), Positives = 64/149 (42%), Gaps = 24/149 (16%)
Query 6 HLRPRTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTLLPE 65
H+ R + M + ++RS + F +RG+ + P + + EG +F +
Sbjct 245 HMMIRGENMSKIFKVRSTVIHCFRNHFFDRGYYEVTPPTLVQTQVEGGSTLFKL------ 298
Query 66 DPKKAVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCALADVYTFGPTFRAENSHTS 125
D+F AYLT S QL +E AL D + ++RAE S T
Sbjct 299 ------------------DYFGEEAYLTQSSQLYLETCIPALGDSFCIAQSYRAEQSRTR 340
Query 126 RHLAEFWMVEPEIAFADLQDDMQLAEQYV 154
RHLAE+ +E E F ++ ++ E V
Sbjct 341 RHLAEYTHIEAECPFMTFENLLERLEDLV 369
> cel:F22D6.3 nrs-1; asparagiNyl tRNA Synthetase family member
(nrs-1); K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=545
Score = 63.9 bits (154), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 44/149 (29%), Positives = 61/149 (40%), Gaps = 24/149 (16%)
Query 6 HLRPRTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTLLPE 65
HL R + + RIR+ + A F G+ + P + + EG +F +
Sbjct 230 HLVIRGENASRILRIRAAATRAMRDHFFAAGYTEVAPPTLVQTQVEGGSTLFGL------ 283
Query 66 DPKKAVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCALADVYTFGPTFRAENSHTS 125
D++ AYLT S QL +E AL DVY ++RAE S T
Sbjct 284 ------------------DYYGEPAYLTQSSQLYLETCNAALGDVYCISQSYRAEKSRTR 325
Query 126 RHLAEFWMVEPEIAFADLQDDMQLAEQYV 154
RHL+E+ VE E AF M E V
Sbjct 326 RHLSEYQHVEAECAFITFDQLMDRIEALV 354
> hsa:1615 DARS, DKFZp781B11202, MGC111579; aspartyl-tRNA synthetase
(EC:6.1.1.12); K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=501
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 43/132 (32%), Positives = 61/132 (46%), Gaps = 25/132 (18%)
Query 10 RTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTLLPEDPKK 69
RT AV R++S + +GF I TP I ++ EG +F+VS
Sbjct 188 RTSTSQAVFRLQSGICHLFRETLINKGFVEIQTPKIISAASEGGANVFTVS--------- 238
Query 70 AVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCA-LADVYTFGPTFRAENSHTSRHL 128
+F +AYL S QL + CA V++ GP FRAE+S+T RHL
Sbjct 239 ---------------YFKNNAYLAQSPQLYKQMCICADFEKVFSIGPVFRAEDSNTHRHL 283
Query 129 AEFWMVEPEIAF 140
EF ++ E+AF
Sbjct 284 TEFVGLDIEMAF 295
> cpv:cgd7_1540 aspartate--tRNA ligase
Length=532
Score = 63.2 bits (152), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 45/134 (33%), Positives = 58/134 (43%), Gaps = 25/134 (18%)
Query 10 RTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTLLPEDPKK 69
RT L A+ RI+S++ F ER F IHTP + E +F V
Sbjct 215 RTYLSQAIFRIQSEVCRLLREFLIEREFIEIHTPKLLPGASESGATVFKV---------- 264
Query 70 AVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCA-LADVYTFGPTFRAENSHTSRHL 128
D+FS A L S QL + C L V+ GP FRAENS+T RHL
Sbjct 265 --------------DYFSNTACLAQSPQLHKQMSICGDLERVFEIGPVFRAENSNTHRHL 310
Query 129 AEFWMVEPEIAFAD 142
EF V+ E+ +
Sbjct 311 CEFVGVDIEMNIEN 324
> dre:568332 nars, fb57a10, si:dkey-274m14.2, wu:fb57a10; asparaginyl-tRNA
synthetase; K01893 asparaginyl-tRNA synthetase
[EC:6.1.1.22]
Length=556
Score = 63.2 bits (152), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 39/149 (26%), Positives = 64/149 (42%), Gaps = 24/149 (16%)
Query 6 HLRPRTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTLLPE 65
H+ R + + + ++RS ++ F RG+ I P + + EG +F +
Sbjct 242 HMMIRGENVSKILKVRSAVTQCFRDHFFSRGYCEITPPTLVQTQVEGGSTLFGL------ 295
Query 66 DPKKAVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCALADVYTFGPTFRAENSHTS 125
++F +AYLT S QL +E AL D + ++RAE S T
Sbjct 296 ------------------NYFGENAYLTQSSQLYLETCIPALGDTFCIAQSYRAEQSRTR 337
Query 126 RHLAEFWMVEPEIAFADLQDDMQLAEQYV 154
RHL+E+ +E E F +D + E V
Sbjct 338 RHLSEYTHIEAECPFMSYEDLLNRLEDLV 366
> tgo:TGME49_002530 aspartyl-tRNA synthetase, putative (EC:6.1.1.12);
K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=773
Score = 62.0 bits (149), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 45/158 (28%), Positives = 67/158 (42%), Gaps = 26/158 (16%)
Query 10 RTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTLLPEDPKK 69
RT + A+ RI+S+ F R F +H+P + EG F++
Sbjct 456 RTPVNQAIFRIQSETLQLFREFLLSRNFVELHSPKLIGGASEGGASCFTLK--------- 506
Query 70 AVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCA-LADVYTFGPTFRAENSHTSRHL 128
+F+R A L S QL + CA V+ GP FRAENS+T RHL
Sbjct 507 ---------------YFNRDACLAQSPQLYKQMAMCADFERVFEIGPVFRAENSNTHRHL 551
Query 129 AEFWMVEPEIAFAD-LQDDMQLAEQYVKFCVKSCLDNC 165
EF ++ E+ F + + + L + KF K + C
Sbjct 552 CEFTGLDLEMTFKEHYSEVLDLLDDLFKFIFKGLSERC 589
> dre:324025 dars, MGC154056, wu:fc17a11, zgc:154056; aspartyl-tRNA
synthetase (EC:6.1.1.12); K01876 aspartyl-tRNA synthetase
[EC:6.1.1.12]
Length=531
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 43/133 (32%), Positives = 61/133 (45%), Gaps = 25/133 (18%)
Query 10 RTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTLLPEDPKK 69
RT A+ R++S + +GF I TP I ++ EG +F+VS
Sbjct 218 RTTTSQAIFRLQSGVCQLFRDTLINKGFVEIQTPKIISAASEGGANVFTVS--------- 268
Query 70 AVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCALAD-VYTFGPTFRAENSHTSRHL 128
+F AYL S QL + CA D V+ GP FRAE+S+T RHL
Sbjct 269 ---------------YFKTSAYLAQSPQLYKQMCICADFDKVFCVGPVFRAEDSNTHRHL 313
Query 129 AEFWMVEPEIAFA 141
EF ++ E+AF+
Sbjct 314 TEFVGLDIEMAFS 326
> xla:380028 dars, MGC53970; aspartyl-tRNA synthetase (EC:6.1.1.12);
K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=530
Score = 58.5 bits (140), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 42/132 (31%), Positives = 58/132 (43%), Gaps = 25/132 (18%)
Query 10 RTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTLLPEDPKK 69
RT AV ++S + +GF I TP I ++ EG +F+VS
Sbjct 217 RTSTSQAVFGLQSGICQLFRDTLTNKGFVEIQTPKIISAASEGGANVFTVS--------- 267
Query 70 AVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCA-LADVYTFGPTFRAENSHTSRHL 128
+F AYL S QL + CA V+ GP FRAE+S+T RHL
Sbjct 268 ---------------YFKTSAYLAQSPQLYKQMCICADFEKVFCIGPVFRAEDSNTHRHL 312
Query 129 AEFWMVEPEIAF 140
EF ++ E+AF
Sbjct 313 TEFVGLDIEMAF 324
> cel:B0464.1 drs-1; aspartyl(D) tRNA Synthetase family member
(drs-1); K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=531
Score = 58.2 bits (139), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 41/132 (31%), Positives = 56/132 (42%), Gaps = 25/132 (18%)
Query 10 RTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTLLPEDPKK 69
RT A+ RI++ + RGF I P I ++ EG +F VS
Sbjct 218 RTPTSHAIFRIQAGICNQFRNILDVRGFVEIMAPKIISAPSEGGANVFEVS--------- 268
Query 70 AVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCA-LADVYTFGPTFRAENSHTSRHL 128
+F AYL S QL + VYT GP FRAE+S+T RH+
Sbjct 269 ---------------YFKGSAYLAQSPQLYKQMAIAGDFEKVYTIGPVFRAEDSNTHRHM 313
Query 129 AEFWMVEPEIAF 140
EF ++ E+AF
Sbjct 314 TEFVGLDLEMAF 325
> ath:AT4G26870 aspartyl-tRNA synthetase, putative / aspartate--tRNA
ligase, putative (EC:6.1.1.12); K01876 aspartyl-tRNA
synthetase [EC:6.1.1.12]
Length=532
Score = 58.2 bits (139), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 40/130 (30%), Positives = 58/130 (44%), Gaps = 25/130 (19%)
Query 10 RTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTLLPEDPKK 69
RT A+ RI+ Q+ +A + Q +GF IHTP + A EG +F +
Sbjct 219 RTPANQAIFRIQCQVQIAFREYLQSKGFLEIHTPKLIAGSSEGGSAVFRL---------- 268
Query 70 AVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCA-LADVYTFGPTFRAENSHTSRHL 128
D+ + A L S QL + C + V+ GP FRAE+S T RHL
Sbjct 269 --------------DYKGQPACLAQSPQLHKQMAICGDMRRVFEVGPVFRAEDSFTHRHL 314
Query 129 AEFWMVEPEI 138
EF ++ E+
Sbjct 315 CEFVGLDVEM 324
> xla:443916 dars, MGC80207; aspartyl-tRNA synthetase (EC:6.1.1.12);
K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=530
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 41/132 (31%), Positives = 59/132 (44%), Gaps = 25/132 (18%)
Query 10 RTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTLLPEDPKK 69
RT +V ++S + ++GF I TP I ++ EG +F+VS
Sbjct 217 RTSTSHSVFGLQSGICQLFRETLTKKGFVEIQTPKIISAASEGGANVFTVS--------- 267
Query 70 AVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCA-LADVYTFGPTFRAENSHTSRHL 128
+F AYL S QL + CA V+ GP FRAE+S+T RHL
Sbjct 268 ---------------YFKTSAYLAQSPQLYKQMCICADFEKVFCIGPVFRAEDSNTHRHL 312
Query 129 AEFWMVEPEIAF 140
EF ++ E+AF
Sbjct 313 TEFVGLDIEMAF 324
> sce:YLL018C DPS1; AspRS; aspartyl-tRNA synthetase (EC:6.1.1.12);
K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=557
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 38/134 (28%), Positives = 57/134 (42%), Gaps = 25/134 (18%)
Query 10 RTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTLLPEDPKK 69
RT A+ RI++ + + + F +HTP + + EG +F V+
Sbjct 240 RTVTNQAIFRIQAGVCELFREYLATKKFTEVHTPKLLGAPSEGGSSVFEVT--------- 290
Query 70 AVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCA-LADVYTFGPTFRAENSHTSRHL 128
+F AYL S Q + A VY GP FRAENS+T RH+
Sbjct 291 ---------------YFKGKAYLAQSPQFNKQQLIVADFERVYEIGPVFRAENSNTHRHM 335
Query 129 AEFWMVEPEIAFAD 142
EF ++ E+AF +
Sbjct 336 TEFTGLDMEMAFEE 349
> bbo:BBOV_IV001690 21.m02784; aspartyl-tRNA synthetase (EC:6.1.1.12);
K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=557
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 41/146 (28%), Positives = 58/146 (39%), Gaps = 32/146 (21%)
Query 10 RTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTLLPEDPKK 69
R L A+ +I+S++ F +R F IH+P + EG +F +
Sbjct 268 RATLNLAIFKIQSEICQQFRNFLLKRDFIEIHSPKLLGGTSEGGCNVFKLK--------- 318
Query 70 AVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCA-LADVYTFGPTFRAENSHTSRHL 128
+F A L S QL + V+ GP FRAENS+T RHL
Sbjct 319 ---------------YFDNDACLAQSPQLYKQLAVVGDFRRVFEIGPVFRAENSNTHRHL 363
Query 129 AEFWMVEPEIAF-------ADLQDDM 147
EF ++ E+ DL DDM
Sbjct 364 CEFVGLDMEMEIRDNYMEVVDLIDDM 389
> pfa:PFA_0145c aspartyl-tRNA synthetase, putative (EC:6.1.1.12);
K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=626
Score = 50.8 bits (120), Expect = 2e-06, Method: Composition-based stats.
Identities = 38/158 (24%), Positives = 64/158 (40%), Gaps = 28/158 (17%)
Query 10 RTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTLLPEDPKK 69
RT ++ ++S + F + F IHTP + EG F +
Sbjct 306 RTYANYSIFSLQSVICHIFRTFLLQHNFVEIHTPKLLGESSEGGANAFKI---------- 355
Query 70 AVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYC--CALADVYTFGPTFRAENSHTSRH 127
++F+++ YL S QL + C V+ GP FRAENS+T RH
Sbjct 356 --------------NYFNQNGYLAQSPQL-YKQMCINSGFDKVFEVGPVFRAENSNTYRH 400
Query 128 LAEFWMVEPEIAFA-DLQDDMQLAEQYVKFCVKSCLDN 164
L E+ ++ E+ + D +++ + K K +N
Sbjct 401 LCEYVSLDIEMTYKFDYMENVHFYDSMFKHIFKELTNN 438
> ath:AT4G31180 aspartyl-tRNA synthetase, putative / aspartate--tRNA
ligase, putative (EC:6.1.1.12); K01876 aspartyl-tRNA
synthetase [EC:6.1.1.12]
Length=558
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 40/158 (25%), Positives = 64/158 (40%), Gaps = 26/158 (16%)
Query 10 RTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTLLPEDPKK 69
RT A+ +++ ++ A + + F IHTP + A EG +F +
Sbjct 245 RTPANQAIFQLQYEVEYAFREKLRFKNFVGIHTPKLMAGSSEGGSAVFRL---------- 294
Query 70 AVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCA-LADVYTFGPTFRAENSHTSRHL 128
++ + A L S QL + C L V+ GP FRAE+S T RHL
Sbjct 295 --------------EYKGQPACLAQSPQLHKQMAICGDLRRVFEVGPVFRAEDSFTHRHL 340
Query 129 AEFWMVEPEIAFAD-LQDDMQLAEQYVKFCVKSCLDNC 165
EF ++ E+ + M L ++ F S + C
Sbjct 341 CEFVGLDVEMEIRKHYSEIMDLVDELFVFIFTSLNERC 378
> tpv:TP03_0489 aspartyl-tRNA synthetase (EC:6.1.1.12); K01876
aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=604
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 39/143 (27%), Positives = 58/143 (40%), Gaps = 26/143 (18%)
Query 15 GAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTLLPEDPKKAVPVN 74
G + +I+S + F F IHTP + EG +F
Sbjct 238 GVIFKIQSAVCQLYREFLLSHDFVEIHTPKLLGGSSEGGSSVFKFK-------------- 283
Query 75 TKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCA-LADVYTFGPTFRAENSHTSRHLAEFWM 133
+F + A L S QL + C L V+ GP FRAENS+T RHL E+
Sbjct 284 ----------YFDQDACLAQSPQLYKQMAICGDLKRVFEIGPVFRAENSNTHRHLCEYVG 333
Query 134 VEPEI-AFADLQDDMQLAEQYVK 155
++ E+ D + + L +Q +K
Sbjct 334 LDMEMEILNDYTEVIDLIDQMLK 356
> bbo:BBOV_IV002630 21.m02971; lysyl-tRNA synthetase (EC:6.1.1.6);
K04567 lysyl-tRNA synthetase, class II [EC:6.1.1.6]
Length=548
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 39/145 (26%), Positives = 52/145 (35%), Gaps = 36/145 (24%)
Query 21 RSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTLLPEDPKKAVPVNTKGEID 80
RS++ T FF +RGF + TP + C GA
Sbjct 182 RSRIVSYTRNFFSQRGFMEVETPTLNVQ-CGGAS-------------------------- 214
Query 81 YAKDFFSRHAYLTVSGQLAVEP-------YCCALADVYTFGPTFRAENSHTSRHLAEFWM 133
A+ F S H L + L V P V+ G FR E + H EF
Sbjct 215 -ARPFISHHNDLDIDLFLRVAPELPLKMIVVGGFEKVFEIGKCFRNEGIDMT-HNPEFTS 272
Query 134 VEPEIAFADLQDDMQLAEQYVKFCV 158
E A+AD D +QL E+Y+ V
Sbjct 273 CEFYWAYADYNDLIQLTEEYLSSLV 297
> eco:b2890 lysS, asuD, ECK2885, herC, JW2858; lysine tRNA synthetase,
constitutive (EC:6.1.1.6); K04567 lysyl-tRNA synthetase,
class II [EC:6.1.1.6]
Length=505
Score = 37.7 bits (86), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 33/151 (21%), Positives = 56/151 (37%), Gaps = 36/151 (23%)
Query 19 RIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTLLPEDPKKAVPVNTKGE 78
++RSQ+ +F RGF + TP++ + +P
Sbjct 185 KVRSQILSGIRQFMVNRGFMEVETPMM-----------------------QVIPGGAA-- 219
Query 79 IDYAKDFFSRHAYLTVSGQLAVEP-------YCCALADVYTFGPTFRAENSHTSRHLAEF 131
A+ F + H L + L + P V+ FR E + RH EF
Sbjct 220 ---ARPFITHHNALDLDMYLRIAPELYLKRLVVGGFERVFEINRNFRNEGI-SVRHNPEF 275
Query 132 WMVEPEIAFADLQDDMQLAEQYVKFCVKSCL 162
M+E +A+AD +D ++L E + + L
Sbjct 276 TMMELYMAYADYKDLIELTESLFRTLAQDIL 306
> tpv:TP04_0470 lysyl-tRNA synthetase; K04567 lysyl-tRNA synthetase,
class II [EC:6.1.1.6]
Length=708
Score = 36.2 bits (82), Expect = 0.045, Method: Composition-based stats.
Identities = 35/134 (26%), Positives = 53/134 (39%), Gaps = 21/134 (15%)
Query 21 RSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTLLPEDPKKAVPVNTKGEID 80
R Q+ RF ERGF + TPI+ E F K+ +N K ++
Sbjct 376 RFQIIAKIRRFLCERGFVEVDTPILNHYTSGDLAEPFETYY------KR---LNEKVKLR 426
Query 81 YAKDFFSRHAYLTVSGQLAVEPYCCALADVYTFGPTFRAENSHTSRHLAEFWMVEPEIAF 140
A +F + ++ + L V+ G FR E S + RH EF M+E
Sbjct 427 IAPEFSIKKIFMGL-----------PLTKVFEIGKCFRNEGS-SLRHSPEFTMIEIYQKM 474
Query 141 ADLQDDMQLAEQYV 154
AD ++L E +
Sbjct 475 ADYNTMIKLLEDLI 488
> sce:YPL104W MSD1, LPG5; Msd1p (EC:6.1.1.12); K01876 aspartyl-tRNA
synthetase [EC:6.1.1.12]
Length=658
Score = 35.8 bits (81), Expect = 0.074, Method: Compositional matrix adjust.
Identities = 42/165 (25%), Positives = 62/165 (37%), Gaps = 23/165 (13%)
Query 3 EIAHLRPRTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTL 62
E +L+ R + RS +S F F + TP++ + EGA E
Sbjct 150 EFRYLQLRNPKYQDFLKKRSSISKEIRNSFNNFDFTEVETPMLFKATPEGAREFL----- 204
Query 63 LPEDPKKAVPVNTKGEIDYAKDFFSRHAYLTVSGQLAVEPYCCALADVYTFGPTFRAENS 122
VP TK D F++ QL + + Y FR E+
Sbjct 205 --------VPTRTKRS-DGKPSFYALDQSPQQYKQLLM---ASGVNKYYQMARCFRDEDL 252
Query 123 HTSRHLAEFWMVEPEIAFADLQDDMQLAEQYV-----KFCVKSCL 162
R EF V+ E+AFA+ +D M++ E+ V KF K L
Sbjct 253 RADRQ-PEFTQVDMEMAFANSEDVMKIIEKTVSGVWSKFSKKRGL 296
> mmu:631033 lysyl-tRNA synthetase-like; K04567 lysyl-tRNA synthetase,
class II [EC:6.1.1.6]
Length=595
Score = 35.0 bits (79), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 37/149 (24%), Positives = 55/149 (36%), Gaps = 36/149 (24%)
Query 20 IRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTLLPEDPKKAVPVNTKGEI 79
+RS++ F E GF I TP++ P AV
Sbjct 244 VRSKIITYIRSFLDELGFLEIETPMMNII------------------PGGAV-------- 277
Query 80 DYAKDFFSRHAYLTVSGQLAVEP-------YCCALADVYTFGPTFRAENSHTSRHLAEFW 132
AK F + H L ++ + + P + VY G FR E + H EF
Sbjct 278 --AKPFITYHNELDMNLYMRIAPELYHKMLVVGGIDRVYEIGRQFRNEGIDLT-HNPEFT 334
Query 133 MVEPEIAFADLQDDMQLAEQYVKFCVKSC 161
E +A+AD D M++ E+ + VKS
Sbjct 335 TCEFYMAYADYHDLMEITEKMLSGMVKSI 363
> mmu:85305 Kars, AA589550, AL024334, AL033315, AL033367, D8Ertd698e,
D8Wsu108e, LysRS, mKIAA0070; lysyl-tRNA synthetase (EC:6.1.1.6);
K04567 lysyl-tRNA synthetase, class II [EC:6.1.1.6]
Length=624
Score = 35.0 bits (79), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 37/149 (24%), Positives = 55/149 (36%), Gaps = 36/149 (24%)
Query 20 IRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTLLPEDPKKAVPVNTKGEI 79
+RS++ F E GF I TP++ P AV
Sbjct 273 VRSKIITYIRSFLDELGFLEIETPMMNII------------------PGGAV-------- 306
Query 80 DYAKDFFSRHAYLTVSGQLAVEP-------YCCALADVYTFGPTFRAENSHTSRHLAEFW 132
AK F + H L ++ + + P + VY G FR E + H EF
Sbjct 307 --AKPFITYHNELDMNLYMRIAPELYHKMLVVGGIDRVYEIGRQFRNEGIDLT-HNPEFT 363
Query 133 MVEPEIAFADLQDDMQLAEQYVKFCVKSC 161
E +A+AD D M++ E+ + VKS
Sbjct 364 TCEFYMAYADYHDLMEITEKMLSGMVKSI 392
> sce:YNR070W PDR18; Pdr18p
Length=1333
Score = 33.9 bits (76), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 36/75 (48%), Gaps = 6/75 (8%)
Query 77 GEIDYAKDFFSRHAYLTVSGQLAVEPYCCALAD---VYTFGPTFRAENSHTSRHLAEFWM 133
G+ AKD+F YL Q E Y A+ D ++ P F + HT+ ++W+
Sbjct 263 GKTTEAKDYFENMGYLCPPRQSTAE-YLTAITDPNGLHEIKPGFEYQVPHTADEFEKYWL 321
Query 134 VEPEIAFADLQDDMQ 148
PE +A L+ ++Q
Sbjct 322 DSPE--YARLKGEIQ 334
> tpv:TP03_0387 lysyl-tRNA synthetase (EC:6.1.1.6); K04567 lysyl-tRNA
synthetase, class II [EC:6.1.1.6]
Length=551
Score = 33.9 bits (76), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 33/143 (23%), Positives = 58/143 (40%), Gaps = 24/143 (16%)
Query 17 VARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTLLPEDPKKAVPVNTK 76
V ++RS++ +F RGF + TP++ + + + F +
Sbjct 214 VMKLRSRIIDYLRKFLTSRGFFEVETPMLKTTSTGASAKPF---------------ITHH 258
Query 77 GEIDYAKDFFSRHAYLTVSGQLAVEPYCC-ALADVYTFGPTFRAENSHTSRHLAEFWMVE 135
E+D D F R ++ +L ++ V+ G FR E + H EF E
Sbjct 259 NELDL--DLFMR-----IAPELPLKLIIIGGFEKVFEIGKCFRNEGIDPT-HNPEFTSCE 310
Query 136 PEIAFADLQDDMQLAEQYVKFCV 158
A+AD D M+L E+++ V
Sbjct 311 FYWAYADYHDLMKLTEEFLSSLV 333
> ath:AT3G13490 OVA5; OVA5 (OVULE ABORTION 5); ATP binding / aminoacyl-tRNA
ligase/ lysine-tRNA ligase/ nucleic acid binding
/ nucleotide binding (EC:6.1.1.6); K04567 lysyl-tRNA synthetase,
class II [EC:6.1.1.6]
Length=602
Score = 33.1 bits (74), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 23/73 (31%), Positives = 32/73 (43%), Gaps = 2/73 (2%)
Query 86 FSRHAYLTVSGQLAVEPYCCA-LADVYTFGPTFRAENSHTSRHLAEFWMVEPEIAFADLQ 144
R YL ++ +L ++ VY G FR E T RH EF +E A++D
Sbjct 298 LGRDLYLRIATELHLKRMLVGGFEKVYEIGRIFRNEGIST-RHNPEFTTIEMYEAYSDYH 356
Query 145 DDMQLAEQYVKFC 157
M +AE V C
Sbjct 357 SMMDMAELIVTQC 369
> sce:YNL073W MSK1; Mitochondrial lysine-tRNA synthetase, required
for import of both aminoacylated and deacylated forms of
tRNA(Lys) into mitochondria and for aminoacylation of mitochondrially
encoded tRNA(Lys) (EC:6.1.1.6); K04567 lysyl-tRNA
synthetase, class II [EC:6.1.1.6]
Length=576
Score = 32.3 bits (72), Expect = 0.72, Method: Composition-based stats.
Identities = 32/140 (22%), Positives = 55/140 (39%), Gaps = 25/140 (17%)
Query 20 IRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTLLPEDPKKAVPVNTKGEI 79
+R+++ +F +R F + TPI+++ + F S+
Sbjct 229 VRARIIKLLRKFLDDRNFVEVETPILSSKSNGAMAKPFITSS------------------ 270
Query 80 DYAKDFFSRHAYLTVSGQLAVEPYCCA-LADVYTFGPTFRAENSHTSRHLAEFWMVEPEI 138
KDF H L ++ +L ++ + L VY G FR E S H AEF +E
Sbjct 271 ---KDF--DHLELRIAPELWLKRLIISGLQKVYEIGKVFRNEGI-DSTHNAEFSTLEFYE 324
Query 139 AFADLQDDMQLAEQYVKFCV 158
+ + D + E KF +
Sbjct 325 TYMSMDDIVTRTEDLFKFLI 344
> eco:b1866 aspS, ECK1867, JW1855, tls; aspartyl-tRNA synthetase
(EC:6.1.1.12); K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=590
Score = 31.2 bits (69), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 19/70 (27%), Positives = 33/70 (47%), Gaps = 7/70 (10%)
Query 6 HLRPRTQLMGAVARIRSQLSMATHRFFQERGFQYIHTPIITASDCEGAGEMFSVSTL--- 62
+L R M + R++++ RF + GF I TP++T + EGA + S +
Sbjct 126 YLDLRRPEMAQRLKTRAKITSLVRRFMDDHGFLDIETPMLTKATPEGARDYLVPSRVHKG 185
Query 63 ----LPEDPK 68
LP+ P+
Sbjct 186 KFYALPQSPQ 195
> hsa:10419 PRMT5, HRMT1L5, IBP72, JBP1, SKB1, SKB1Hs; protein
arginine methyltransferase 5 (EC:2.1.1.125); K02516 protein
arginine N-methyltransferase 5 [EC:2.1.1.125]
Length=620
Score = 29.6 bits (65), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 12/25 (48%), Positives = 16/25 (64%), Gaps = 0/25 (0%)
Query 123 HTSRHLAEFWMVEPEIAFADLQDDM 147
HT H + FWM P +A DL+DD+
Sbjct 126 HTGHHSSMFWMRVPLVAPEDLRDDI 150
Lambda K H
0.324 0.135 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4027755848
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40