bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0011_orf1
Length=306
Score E
Sequences producing significant alignments: (Bits) Value
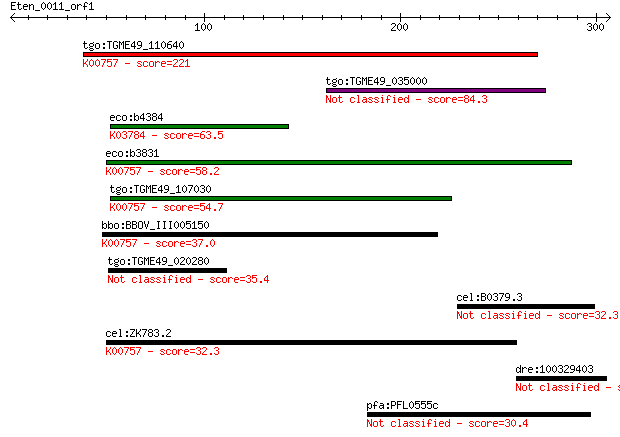
tgo:TGME49_110640 uridine phosphorylase, putative (EC:2.4.2.1)... 221 2e-57
tgo:TGME49_035000 hypothetical protein 84.3 5e-16
eco:b4384 deoD, ECK4376, JW4347, pup; purine-nucleoside phosph... 63.5 8e-10
eco:b3831 udp, ECK3825, JW3808; uridine phosphorylase (EC:2.4.... 58.2 4e-08
tgo:TGME49_107030 purine nucleoside phosphorylase (EC:2.4.2.3)... 54.7 4e-07
bbo:BBOV_III005150 17.m07460; hypothetical protein; K00757 uri... 37.0 0.093
tgo:TGME49_020280 SCP-like domain-containing protein 35.4 0.26
cel:B0379.3 mut-16; MUTator family member (mut-16) 32.3 2.4
cel:ZK783.2 upp-1; Uridine PhosPhorylase family member (upp-1)... 32.3 2.4
dre:100329403 hypothetical protein LOC100329403 31.2 5.4
pfa:PFL0555c conserved Plasmodium protein 30.4 8.0
> tgo:TGME49_110640 uridine phosphorylase, putative (EC:2.4.2.1);
K00757 uridine phosphorylase [EC:2.4.2.3]
Length=303
Score = 221 bits (564), Expect = 2e-57, Method: Compositional matrix adjust.
Identities = 111/236 (47%), Positives = 148/236 (62%), Gaps = 4/236 (1%)
Query 38 KEAEIFLNEDGSLYHLGVKSGELHPRILTVGDRERAQMIAEAFLEQVREFEKSRNFRTFS 97
K IFL DG YHLGVK G+L I+TVG +RA+++A FL V E SR F TF+
Sbjct 5 KGKNIFLTPDGRTYHLGVKKGDLASLIVTVGCEQRARVLAAKFLTDVTEVSSSRQFYTFT 64
Query 98 GLY--KGRGVSVVCIGMGAPNMDFLVRESSFLFQQKGLAFVRLGTCGIFKAGCSPGTLCI 155
G Y G+ +S+VCIGMGAP DF +RE+SF+ + + LA VR+G+CGI PGTL +
Sbjct 65 GKYIKGGQRMSIVCIGMGAPMADFFIREASFVLEGEPLAVVRIGSCGIVDYATRPGTLML 124
Query 156 SNSIYYCQRNYAHFDGSAAGAPGVHTPEG--PYLISGPVKGDEELLNKIEENVKRKNLAY 213
S+ YC RNY HFDG AA PYL++ PVK D+ L + IE N+ + A
Sbjct 125 SDESMYCYRNYCHFDGEAALGNKRECGRDPRPYLLTAPVKADKRLTDLIEANLNAQGAAV 184
Query 214 LRGLGITAETFYSCQGRRFPGFGDENEKLVEDFVRLGIDSCEMESHQLYHLCSERE 269
RGL +AETF++CQGR P + D N L++D LG+ S EME+HQ++HL +R+
Sbjct 185 KRGLNCSAETFFACQGRELPLWNDNNANLLKDMTDLGVVSLEMETHQIFHLMHQRQ 240
> tgo:TGME49_035000 hypothetical protein
Length=161
Score = 84.3 bits (207), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 51/119 (42%), Positives = 67/119 (56%), Gaps = 12/119 (10%)
Query 162 CQRNYAHFDGS---AAGAPGVHTPEGPYLISGPVKGDEELLNKIEENVKRK----NLAYL 214
C +Y FDGS A GV PY+I+ P D EL N+I EN+ + +L +L
Sbjct 3 CYNDYTFFDGSMEDGACLKGV----SPYVITRPCAPDHELSNRIVENISKAVDDPSLLHL 58
Query 215 RGLGITAETFYSCQGRRFPGFGDENEKLVEDFVRLGIDSCEMESHQLYHLCSEREKERK 273
GL +AE FY+CQG F D NEKL++ + G+DS +MESHQL HL + R K K
Sbjct 59 -GLHASAENFYACQGHVDNLFNDRNEKLIDQLLSHGVDSMDMESHQLLHLANRRFKPVK 116
> eco:b4384 deoD, ECK4376, JW4347, pup; purine-nucleoside phosphorylase
(EC:2.4.2.1); K03784 purine-nucleoside phosphorylase
[EC:2.4.2.1]
Length=239
Score = 63.5 bits (153), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 35/91 (38%), Positives = 49/91 (53%), Gaps = 2/91 (2%)
Query 52 HLGVKSGELHPRILTVGDRERAQMIAEAFLEQVREFEKSRNFRTFSGLYKGRGVSVVCIG 111
H+ + G+ +L GD RA+ IAE FLE RE R F+G YKGR +SV+ G
Sbjct 5 HINAEMGDFADVVLMPGDPLRAKYIAETFLEDAREVNNVRGMLGFTGTYKGRKISVMGHG 64
Query 112 MGAPNMDFLVRESSFLFQQKGLAFVRLGTCG 142
MG P+ +E F K + +R+G+CG
Sbjct 65 MGIPSCSIYTKELITDFGVKKI--IRVGSCG 93
> eco:b3831 udp, ECK3825, JW3808; uridine phosphorylase (EC:2.4.2.3);
K00757 uridine phosphorylase [EC:2.4.2.3]
Length=253
Score = 58.2 bits (139), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 58/255 (22%), Positives = 103/255 (40%), Gaps = 39/255 (15%)
Query 50 LYHLGVKSGELHPRILTV--GDRERAQMIAEAFLEQVREFEKSRNFRTFSGLYKGRGVSV 107
++HLG+ +L L + GD +R + IA A +++ + R F T+ G+ V V
Sbjct 6 VFHLGLTKNDLQGATLAIVPGDPDRVEKIA-ALMDKPVKLASHREFTTWRAELDGKPVIV 64
Query 108 VCIGMGAPNMDFLVRESSFLFQQKGL-AFVRLGTCGIFKAGCSPGTLCISNSIYYCQRNY 166
G+G P+ V E Q G+ F+R+GT G + + G + ++ +
Sbjct 65 CSTGIGGPSTSIAVEE----LAQLGIRTFLRIGTTGAIQPHINVGDVLVTTASVRLDGAS 120
Query 167 AHFDGSAAGAPGVHTPEGPYLISGPVKGDEELLNKIEENVKRKNLAYLRGLGITAETFYS 226
HF AP + P D E + E K G+ +++TFY
Sbjct 121 LHF------AP----------LEFPAVADFECTTALVEAAKSIGATTHVGVTASSDTFYP 164
Query 227 CQGRRFPGFGDENEKL---VEDFVRLGIDSCEMESHQLYHLCSER------------EKE 271
Q R G +E++ +G+ + EMES L +C+ + +
Sbjct 165 GQERYDTYSGRVVRHFKGSMEEWQAMGVMNYEMESATLLTMCASQGLRAGMVAGVIVNRT 224
Query 272 RKRKENEKEMKEKET 286
++ N + MK+ E+
Sbjct 225 QQEIPNAETMKQTES 239
> tgo:TGME49_107030 purine nucleoside phosphorylase (EC:2.4.2.3);
K00757 uridine phosphorylase [EC:2.4.2.3]
Length=247
Score = 54.7 bits (130), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 40/175 (22%), Positives = 79/175 (45%), Gaps = 21/175 (12%)
Query 52 HLGVKSGELHPRILTVGDRERAQMIAEAFLEQVREFEKSRNFRTFSGLYKGRGVSVVCIG 111
H+ ++ ++ P ++ VGD R + +A E+ +E +R +R+F +Y + ++V+ G
Sbjct 9 HIRLRKEDVEPVVIIVGDPARTEEVAN-MCEKKQELAYNREYRSFRVVYDSQPITVISHG 67
Query 112 MGAPNMDFLVRESSFLFQQKGLAFVRLGTCGIFKA-GCSPGTLCISNSIYYCQRNYAHFD 170
+G P + E ++L + +R GTCG K G +C++ YA +
Sbjct 68 IGCPGTSIAIEELAYLGAK---VIIRAGTCGSLKPKTLKQGDVCVT---------YAAVN 115
Query 171 GSAAGAPGVHTPEGPYLISGPVKGDEELLNKIEENVKRKNLAYLRGLGITAETFY 225
+ G PEG ++ P + + + K + G+G+T + FY
Sbjct 116 ET--GLISNILPEGFPCVATP-----HVYQALMDAAKELGIEAASGIGVTQDYFY 163
> bbo:BBOV_III005150 17.m07460; hypothetical protein; K00757 uridine
phosphorylase [EC:2.4.2.3]
Length=260
Score = 37.0 bits (84), Expect = 0.093, Method: Compositional matrix adjust.
Identities = 40/174 (22%), Positives = 73/174 (41%), Gaps = 13/174 (7%)
Query 48 GSLYHLGVKSGELHPRILTVGDRERAQMIAEAFLEQVREFEKSRNFRTFSGLYKGRGVSV 107
G + +GV +HP + VGD + +M+A Q F + ++ + + G+
Sbjct 10 GIMNKIGVPLDRIHPVAIVVGDPKWLEMLASLATRQ-EFFARYKSLSSMELEFNGQTFFA 68
Query 108 VCIGMGAPNMDFLVRESSFLFQQKGLAFVRLGTCGIFKAGCSPGTLCISNSIYYCQRNYA 167
+ G GA N+ L+ E + +F K + + T + LCI+ + C+ +YA
Sbjct 69 LSFGFGATNLHRLLTELA-VFGVKCVILIT-NTFSLLPGRVPANALCITYAA--CRGDYA 124
Query 168 HFDGSAAGAPGVHTPEGPYLISGPVKGDEELLNKIEENVKR---KNLAYLRGLG 218
P + P+ + K EEL K++ ++ R + AY LG
Sbjct 125 STIEVDLAYPAIAHPDATRALR---KAAEEL--KLDYHLTRTLTNDYAYNSKLG 173
> tgo:TGME49_020280 SCP-like domain-containing protein
Length=434
Score = 35.4 bits (80), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 24/66 (36%), Positives = 34/66 (51%), Gaps = 10/66 (15%)
Query 51 YHLGVKSGELHPRI--LTVGDRERAQMIAEAFL----EQVREFEKSRNFRTFSGLYKGRG 104
YH + +L PR+ LT GD+ERA +AF+ E V E ++ RN G+ G
Sbjct 62 YHDAKRGRQLSPRLSELTTGDKERASQRLQAFIDNGCEDVVEVDECRN----KGINVFDG 117
Query 105 VSVVCI 110
SV C+
Sbjct 118 TSVSCV 123
> cel:B0379.3 mut-16; MUTator family member (mut-16)
Length=1054
Score = 32.3 bits (72), Expect = 2.4, Method: Composition-based stats.
Identities = 23/74 (31%), Positives = 39/74 (52%), Gaps = 6/74 (8%)
Query 229 GRRFPGFGDENEKLVEDFVRLGIDSCEMESHQLYHLCSEREKERKRKENEKEMKEKETKD 288
G R P +GD E+ V+D DS E+ S Y +KE KR+ + ++K+K+ ++
Sbjct 501 GNRGPSYGDRGER-VQDVGDTTSDS-EITSEGSYSDEDPEQKEIKRQRRKDKLKKKQERE 558
Query 289 ----NSNSESKQGP 298
+++SKQ P
Sbjct 559 LRSREKHTKSKQQP 572
> cel:ZK783.2 upp-1; Uridine PhosPhorylase family member (upp-1);
K00757 uridine phosphorylase [EC:2.4.2.3]
Length=295
Score = 32.3 bits (72), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 58/229 (25%), Positives = 84/229 (36%), Gaps = 37/229 (16%)
Query 50 LYHLGVKSGELH-PRIL-------TVGDRERAQMIAEAFLEQVR-----EFEKSRNFRTF 96
LYH G L P + T G R ++ AE F ++ +S F
Sbjct 26 LYHFGFGVDTLDIPAVFGDTKFVCTGGSPGRFKLYAEWFAKEANIPCSENLSRSDRFV-- 83
Query 97 SGLYKGRGVSVVCIGMGAPNMDFLVRESSFLFQQKGL---AFVRLGTCGIFKAGCSPGTL 153
+YK V + GMG P++ ++ ES L G+ F+RLGT G +
Sbjct 84 --IYKTGPVCWINHGMGTPSLSIMLVESFKLMHHAGVKNPTFIRLGTSGGVGVPPGTVVV 141
Query 154 CISNSIYYCQRNYAHFDGSAAGAPGVHTPEGPYLISGPVKGDEELLNKIEENVKRKNLAY 213
+ G V G I P + D L + E K K++
Sbjct 142 STEAM------------NAELGDTYVQIIAGKR-IERPTQLDAALREALCEVGKEKSIPV 188
Query 214 LRGLGITAETFYSCQGRRFPGFGDENEKLVEDFVR----LGIDSCEMES 258
G + A+ FY Q R F D E+ F+R LG+ + EMES
Sbjct 189 ETGKTMCADDFYEGQMRLDGYFCDYEEEDKYAFLRKLNALGVRNIEMES 237
> dre:100329403 hypothetical protein LOC100329403
Length=409
Score = 31.2 bits (69), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 29/49 (59%), Gaps = 7/49 (14%)
Query 259 HQLYH---LCSEREKERKRKENEKEMKEKETKDNSNSESKQGPTRAASI 304
+++YH LC+E ++E +R+EN+++ E K N SKQ PT I
Sbjct 320 NRVYHALALCNEIQRENERRENQRKEDEVWVKQN----SKQCPTCGVKI 364
> pfa:PFL0555c conserved Plasmodium protein
Length=1831
Score = 30.4 bits (67), Expect = 8.0, Method: Composition-based stats.
Identities = 31/120 (25%), Positives = 57/120 (47%), Gaps = 10/120 (8%)
Query 183 EGPYLISGPVKGDEELLNKIEEN-VKRKNLAYLRGLGITAETFYSCQGRRFPGFGDEN-- 239
E Y+ S + E+ N EN VK L L+ L + E + + F + +EN
Sbjct 979 ETNYIKSSCHNSEVEIYNSTRENNVKNNKLEKLKHLKVKFEDDH--KRINFKNYEEENIK 1036
Query 240 ---EKLVEDFVRLGIDSCEMESHQLYHLCSEREKERKRKENEKEMKEKETKDNSNSESKQ 296
K +E F RL + +E + YH + +++ +K++ K K+ +T++N N +K+
Sbjct 1037 GHKNKYIE-FYRLDENESNVECTK-YHTNKKDDEKNYKKKSTKIQKDIKTQENKNDITKE 1094
Lambda K H
0.319 0.136 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 12347680184
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40