bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0072_orf2
Length=382
Score E
Sequences producing significant alignments: (Bits) Value
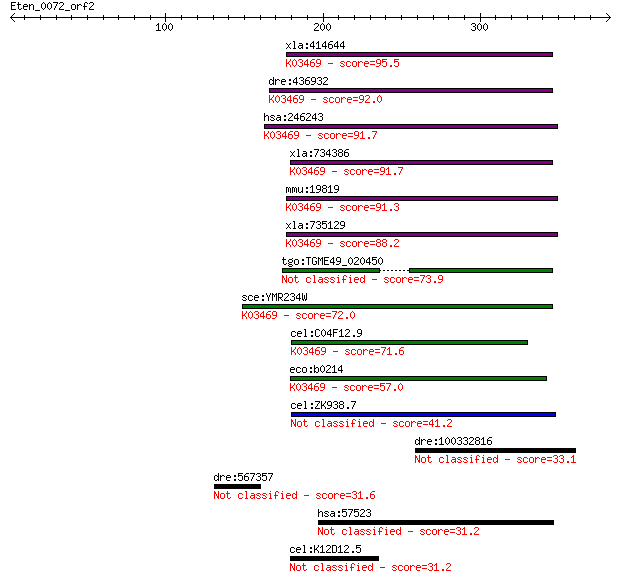
xla:414644 hypothetical protein MGC81203; K03469 ribonuclease ... 95.5 3e-19
dre:436932 rnaseh1, zgc:91971; ribonuclease H1 (EC:3.1.26.4); ... 92.0 3e-18
hsa:246243 RNASEH1, H1RNA, RNH1; ribonuclease H1 (EC:3.1.26.4)... 91.7 4e-18
xla:734386 hypothetical protein MGC115038; K03469 ribonuclease... 91.7 4e-18
mmu:19819 Rnaseh1; ribonuclease H1 (EC:3.1.26.4); K03469 ribon... 91.3 6e-18
xla:735129 rnaseh1, MGC115609, h1rna, rnh1; ribonuclease H1; K... 88.2 5e-17
tgo:TGME49_020450 ribonuclease H, putative (EC:3.1.26.4) 73.9 1e-12
sce:YMR234W RNH1; Ribonuclease H1; able to bind double-strande... 72.0 4e-12
cel:C04F12.9 rnh-1.3; RNase H family member (rnh-1.3); K03469 ... 71.6 5e-12
eco:b0214 rnhA, cer, dasF, ECK0214, herA, JW0204, rnh, sdrA, s... 57.0 1e-07
cel:ZK938.7 rnh-1.2; RNase H family member (rnh-1.2) 41.2 0.007
dre:100332816 RETRotransposon-like family member (retr-1)-like 33.1 2.1
dre:567357 Sushi, von Willebrand factor type A, EGF and pentra... 31.6 5.0
hsa:57523 NYNRIN, CGIN1, FLJ11811, KIAA1305; NYN domain and re... 31.2 6.5
cel:K12D12.5 hypothetical protein 31.2 6.6
> xla:414644 hypothetical protein MGC81203; K03469 ribonuclease
HI [EC:3.1.26.4]
Length=300
Score = 95.5 bits (236), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 59/169 (34%), Positives = 86/169 (50%), Gaps = 25/169 (14%)
Query 177 DVVEIYADGACPSNGRVDARAGIGVFFGTDDRRYLFMPLVVGPQTYQRAELASILAALLR 236
D +Y DG C NGRV ARAGIGV++G L L G QT QRAE+ + AL
Sbjct 150 DAAVVYTDGCCSGNGRVKARAGIGVYWGQGHPLNLAEKLE-GRQTNQRAEIQAACRAL-- 206
Query 237 FVDYDDDSAKQQPSGRSEVRLVVISDSSYAVNCLGPWAARWCRNGWKSTSGKPVKNADLI 296
+Q ++ +LV+ +DS + +N + W W RNGWK ++GK V N +
Sbjct 207 ----------EQAKAQNLTKLVLYTDSMFTINGITKWIHSWKRNGWKLSTGKNVINREDF 256
Query 297 QAILLLCDKRRKTAPSGATWRGSVEYEYIRGHSGVYGNEMADKLAVQGA 345
+ + + T +++ +I GH+G GNE ADKL+ +GA
Sbjct 257 ENLEKM------------TQGLDIKWMHIPGHAGFAGNEAADKLSREGA 293
> dre:436932 rnaseh1, zgc:91971; ribonuclease H1 (EC:3.1.26.4);
K03469 ribonuclease HI [EC:3.1.26.4]
Length=276
Score = 92.0 bits (227), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 64/186 (34%), Positives = 88/186 (47%), Gaps = 31/186 (16%)
Query 166 PQPETATPMKP------DVVEIYADGACPSNGRVDARAGIGVFFGTDDRRYLFMPLVVGP 219
PQP+T T D V +Y DG C NG+ ARAGIGV++G D R + L G
Sbjct 111 PQPKTGTTSSDGFTYMGDAVVVYTDGCCSGNGKHGARAGIGVYWGRDHPRNVAERL-PGR 169
Query 220 QTYQRAELASILAALLRFVDYDDDSAKQQPSGRSEVRLVVISDSSYAVNCLGPWAARWCR 279
QT QRAEL + AL +Q + ++V+ +DS + +N + W W
Sbjct 170 QTNQRAELQAACKAL------------EQAKEMNFKKVVLYTDSKFTINGVTSWVKTWKS 217
Query 280 NGWKSTSGKPVKNADLIQAILLLCDKRRKTAPSGATWRGSVEYEYIRGHSGVYGNEMADK 339
NGW+ SG + N D Q + L A W +I GH+G GNE AD+
Sbjct 218 NGWRLKSGGVIINKDDFQQLDKL------NAELDVVWM------HIPGHAGYTGNEEADR 265
Query 340 LAVQGA 345
L+ +GA
Sbjct 266 LSREGA 271
> hsa:246243 RNASEH1, H1RNA, RNH1; ribonuclease H1 (EC:3.1.26.4);
K03469 ribonuclease HI [EC:3.1.26.4]
Length=286
Score = 91.7 bits (226), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 66/195 (33%), Positives = 97/195 (49%), Gaps = 40/195 (20%)
Query 163 SVHPQP----ETATPMKPDVVEIYADGACPSNGRVDARAGIGVFFGTDDRRYLFMPLVVG 218
SV P P +T + M D V +Y DG C SNGR RAGIGV++G PL VG
Sbjct 121 SVEPAPPVSRDTFSYMG-DFVVVYTDGCCSSNGRRRPRAGIGVYWGPGH------PLNVG 173
Query 219 -----PQTYQRAELASILAALLRFVDYDDDSAKQQPSGRSEVRLVVISDSSYAVNCLGPW 273
QT QRAE+ + A+ +Q ++ +LV+ +DS + +N + W
Sbjct 174 IRLPGRQTNQRAEIHAACKAI------------EQAKTQNINKLVLYTDSMFTINGITNW 221
Query 274 AARWCRNGWKSTSGKPVKNADLIQAILLLCDKRRKTAPSGATWRGSVEYEYIRGHSGVYG 333
W +NGWK+++GK V N + A+ L T +++ ++ GHSG G
Sbjct 222 VQGWKKNGWKTSAGKEVINKEDFVALERL------------TQGMDIQWMHVPGHSGFIG 269
Query 334 NEMADKLAVQGANSA 348
NE AD+LA +GA +
Sbjct 270 NEEADRLAREGAKQS 284
> xla:734386 hypothetical protein MGC115038; K03469 ribonuclease
HI [EC:3.1.26.4]
Length=204
Score = 91.7 bits (226), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 54/167 (32%), Positives = 80/167 (47%), Gaps = 25/167 (14%)
Query 179 VEIYADGACPSNGRVDARAGIGVFFGTDDRRYLFMPLVVGPQTYQRAELASILAALLRFV 238
E+Y DG C NG+ A GIGV++G +D R + L G QT QRAE+ +
Sbjct 61 AEVYTDGCCSRNGQYGANGGIGVYWGPNDSRNVSARL-EGRQTNQRAEIEAAY------- 112
Query 239 DYDDDSAKQQPSGRSEVRLVVISDSSYAVNCLGPWAARWCRNGWKSTSGKPVKNADLIQA 298
+A +Q + L + +DS + ++ + W RW N WK+ G+ V N Q
Sbjct 113 -----TAAKQARNDNVTHLEINTDSKFTIDGMTKWVPRWKENNWKTIDGRDVINKQDFQK 167
Query 299 ILLLCDKRRKTAPSGATWRGSVEYEYIRGHSGVYGNEMADKLAVQGA 345
+ C+ V++ Y+ GHSG GNEMAD+LA G+
Sbjct 168 LDKACENM------------DVKWNYVPGHSGNAGNEMADQLAKAGS 202
> mmu:19819 Rnaseh1; ribonuclease H1 (EC:3.1.26.4); K03469 ribonuclease
HI [EC:3.1.26.4]
Length=285
Score = 91.3 bits (225), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 59/177 (33%), Positives = 89/177 (50%), Gaps = 35/177 (19%)
Query 177 DVVEIYADGACPSNGRVDARAGIGVFFGTDDRRYLFMPLVVG-----PQTYQRAELASIL 231
+ V +Y DG C SNGR ARAGIGV++G PL VG QT QRAE+ +
Sbjct 137 ESVVVYTDGCCSSNGRKRARAGIGVYWGPGH------PLNVGIRLPGRQTNQRAEIHAAC 190
Query 232 AALLRFVDYDDDSAKQQPSGRSEVRLVVISDSSYAVNCLGPWAARWCRNGWKSTSGKPVK 291
A++ Q ++ +LV+ +DS + +N + W W +NGW++++GK V
Sbjct 191 KAIM------------QAKAQNISKLVLYTDSMFTINGITNWVQGWKKNGWRTSTGKDVI 238
Query 292 NADLIQAILLLCDKRRKTAPSGATWRGSVEYEYIRGHSGVYGNEMADKLAVQGANSA 348
N + + L T +++ +I GHSG GNE AD+LA +GA +
Sbjct 239 NKEDFMELDEL------------TQGMDIQWMHIPGHSGFVGNEEADRLAREGAKQS 283
> xla:735129 rnaseh1, MGC115609, h1rna, rnh1; ribonuclease H1;
K03469 ribonuclease HI [EC:3.1.26.4]
Length=292
Score = 88.2 bits (217), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 57/173 (32%), Positives = 86/173 (49%), Gaps = 27/173 (15%)
Query 177 DVVEIYADGACPSNGRVDARAGIGVFFGTDDRRYLFMPLVVGPQTYQRAELASILAALLR 236
D +Y DG C NGR+ A+AGIGV++G L L G QT QRAE+ + AL
Sbjct 143 DSAVVYTDGCCSQNGRLKAQAGIGVYWGPGHPLNLAERLE-GRQTNQRAEIQAACRAL-- 199
Query 237 FVDYDDDSAKQQPSGRSEVRLVVISDSSYAVNCLGPWAARWCRNGWKSTSGKPVKNADLI 296
+Q ++ +LV+ +DS + +N + W W R GWK ++G+ V N +
Sbjct 200 ----------EQAQAKNITKLVLYTDSMFTINGITKWIHSWKRKGWKLSNGQNVINREDF 249
Query 297 QAILLLCDKRRKTAPSGATWRG-SVEYEYIRGHSGVYGNEMADKLAVQGANSA 348
+ + L RG + + +I GH+G GNE ADKL+ +GA +
Sbjct 250 EKLDKLT-------------RGLDINWMHIPGHAGFEGNEAADKLSREGAQKS 289
> tgo:TGME49_020450 ribonuclease H, putative (EC:3.1.26.4)
Length=657
Score = 73.9 bits (180), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 42/120 (35%), Positives = 57/120 (47%), Gaps = 29/120 (24%)
Query 255 VRLVVISDSSYAVNCLGPWAARWCRNGWKSTSGKPVKNADLIQAILLLCDKRR------- 307
+ + + SDS YAV C+ W W NGW++++G V+N DLI I L R+
Sbjct 526 LEMRICSDSVYAVRCVTEWVTAWKANGWRTSAGTEVRNRDLIAKIHELLQSRKLGDARFS 585
Query 308 --------------------KTAPSGATW--RGSVEYEYIRGHSGVYGNEMADKLAVQGA 345
AP W RG++E+ I+GHSG YGNE AD+LA GA
Sbjct 586 SETPELKVERERDSGMSRKEHKAPIQGGWGGRGTIEFVLIKGHSGNYGNEKADQLACLGA 645
Score = 64.7 bits (156), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 35/65 (53%), Positives = 44/65 (67%), Gaps = 3/65 (4%)
Query 174 MKPD--VVEIYADGACPSNGR-VDARAGIGVFFGTDDRRYLFMPLVVGPQTYQRAELASI 230
M+ D ++ IY DGAC SNGR +A+AG+GVFFG D R + L PQT QRAEL +I
Sbjct 351 MRDDARILYIYTDGACRSNGRGKEAKAGVGVFFGDGDPRNVSRRLTGQPQTNQRAELQAI 410
Query 231 LAALL 235
L AL+
Sbjct 411 LDALV 415
> sce:YMR234W RNH1; Ribonuclease H1; able to bind double-stranded
RNAs and RNA-DNA hybrids; associates with RNAse polymerase
I; the homolog of mammalian RNAse HII (the S. cerevisiae
homolog of mammalian RNAse HI is RNH201) (EC:3.1.26.4); K03469
ribonuclease HI [EC:3.1.26.4]
Length=348
Score = 72.0 bits (175), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 55/200 (27%), Positives = 90/200 (45%), Gaps = 15/200 (7%)
Query 149 AADHHRLLNCSPGASVHPQPETATPMKPDVVEIYADGACPSNGRVDARAGIGVFFGTDDR 208
+A ++L+N S + ++ M + +Y DG+ NG +RAG G +F
Sbjct 158 SAHDYKLMNISKESFESKYKLSSNTMYNKSMNVYCDGSSFGNGTSSSRAGYGAYFEGAPE 217
Query 209 RYLFMPLVVGPQTYQRAELASILAALLRFVDYDDDSAKQQPSGRSEVRLVVISDSSYAVN 268
+ PL+ G QT RAE+ ++ AL + + + + + +V + +DS Y
Sbjct 218 ENISEPLLSGAQTNNRAEIEAVSEALKKIWE-------KLTNEKEKVNYQIKTDSEYVTK 270
Query 269 CLGPWAARWCRNGWKSTSGKPVKNADLIQAILLLCDKRRKTAPSGATW---RGSVEYEYI 325
L R+ K G P N+DLI ++ K +K G + E++
Sbjct 271 LLND---RYMTYDNKKLEGLP--NSDLIVPLVQRFVKVKKYYELNKECFKNNGKFQIEWV 325
Query 326 RGHSGVYGNEMADKLAVQGA 345
+GH G GNEMAD LA +GA
Sbjct 326 KGHDGDPGNEMADFLAKKGA 345
> cel:C04F12.9 rnh-1.3; RNase H family member (rnh-1.3); K03469
ribonuclease HI [EC:3.1.26.4]
Length=139
Score = 71.6 bits (174), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 51/150 (34%), Positives = 72/150 (48%), Gaps = 19/150 (12%)
Query 180 EIYADGACPSNGRVDARAGIGVFFGTDDRRYLFMPLVVGPQTYQRAELASILAALLRFVD 239
+ Y DG+ NG+ +R G GV +D + + GPQT R EL +I A D
Sbjct 4 DFYTDGSALGNGQQGSRGGYGVHVPSDTSKNVSGSYPHGPQTNNRYELEAIKHATQMARD 63
Query 240 YDDDSAKQQPSGRSEVRLVVISDSSYAVNCLGPWAARWCRNGWKSTSGKPVKNADLIQAI 299
D S VR + +DS A + + W W NG+K++SG+ VKN DLI+ I
Sbjct 64 VPD----------SHVR--IHTDSKNAKDSVTKWNDNWKSNGYKTSSGQDVKNQDLIRDI 111
Query 300 LLLCDKRRKTAPSGATWRGSVEYEYIRGHS 329
+ SG T VE+E++RGHS
Sbjct 112 DRNV---QALKHSGKT----VEFEHVRGHS 134
> eco:b0214 rnhA, cer, dasF, ECK0214, herA, JW0204, rnh, sdrA,
sin; ribonuclease HI, degrades RNA of DNA-RNA hybrids (EC:3.1.26.4);
K03469 ribonuclease HI [EC:3.1.26.4]
Length=155
Score = 57.0 bits (136), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 43/163 (26%), Positives = 66/163 (40%), Gaps = 30/163 (18%)
Query 179 VEIYADGACPSNGRVDARAGIGVFFGTDDRRYLFMPLVVGPQTYQRAELASILAALLRFV 238
VEI+ DG+C N G G R F T R EL + + AL
Sbjct 5 VEIFTDGSCLGN---PGPGGYGAILRYRGREKTFSAGYT-RTTNNRMELMAAIVAL---- 56
Query 239 DYDDDSAKQQPSGRSEVRLVVISDSSYAVNCLGPWAARWCRNGWKSTSGKPVKNADLIQA 298
+ + +++ +DS Y + W W + GWK+ KPVKN DL Q
Sbjct 57 ----------EALKEHCEVILSTDSQYVRQGITQWIHNWKKRGWKTADKKPVKNVDLWQR 106
Query 299 ILLLCDKRRKTAPSGATWRGSVEYEYIRGHSGVYGNEMADKLA 341
+ A + +++E+++GH+G NE D+LA
Sbjct 107 L------------DAALGQHQIKWEWVKGHAGHPENERCDELA 137
> cel:ZK938.7 rnh-1.2; RNase H family member (rnh-1.2)
Length=404
Score = 41.2 bits (95), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 49/171 (28%), Positives = 69/171 (40%), Gaps = 12/171 (7%)
Query 180 EIYADGACPSNGRVDARAGIGVFFGTDDR--RYLFMPLVVGPQTYQRAELASILAALLRF 237
E+Y DG+ +NG+ AR G + F D Y FM VG QT EL +I A
Sbjct 7 EVYTDGSTVNNGKRGARGGWAIVFPFDRSLDEYDFMK--VGKQTNNVYELTAIYEATEVV 64
Query 238 VDYDDDSAKQQPSGRSEVRLVVISDSSYAVNCLGPWAARWCRNGWKSTSGKPVKNADLIQ 297
+D Q+ + + SS + C + NG ST G + +
Sbjct 65 RCWDSFKCNQKLEFVRLKNTITPTTSSILIPCTRRTVS---PNG--STDGTNMGGLRITP 119
Query 298 AILLLCDKRRKTAPSGATWRG-SVEYEYIRGHSGVYGNEMADKLAVQGANS 347
I L R PS A V+ EY++GH + N AD+LA + S
Sbjct 120 GI--LSKISRSFVPSTAISESLDVKIEYVKGHHTNFFNCEADRLAKKACRS 168
> dre:100332816 RETRotransposon-like family member (retr-1)-like
Length=1236
Score = 33.1 bits (74), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 29/111 (26%), Positives = 45/111 (40%), Gaps = 20/111 (18%)
Query 259 VISDSSYAVNCLGPWAARWCRNGWKSTSGKPVKNADLIQAILLLCDKRRKTAPSGATWRG 318
+ +DS YA + A W G+ + GKP+ ++ L+ ++ A
Sbjct 796 IYTDSRYAFGVAHDFGAIWASRGFVAADGKPISHSSLVMDLI-----------KAARLPR 844
Query 319 SVEYEYIRGHSG-----VYGNEMAD---KLAVQ-GANSAWPAHSLASGAGF 360
S+ RGHS GN AD KLA Q G +W S+ S + +
Sbjct 845 SLAIIKTRGHSTFDTEEAKGNNFADTQAKLAAQSGKQPSWLNASMVSTSPY 895
> dre:567357 Sushi, von Willebrand factor type A, EGF and pentraxin
domain-containing protein 1-like
Length=3644
Score = 31.6 bits (70), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 15/33 (45%), Positives = 18/33 (54%), Gaps = 4/33 (12%)
Query 131 AQPKETWGGAEPSGRELNAAD----HHRLLNCS 159
QP TW G +PS R + D HH +LNCS
Sbjct 414 CQPDGTWSGVQPSCRIRSCPDLSPPHHGMLNCS 446
> hsa:57523 NYNRIN, CGIN1, FLJ11811, KIAA1305; NYN domain and
retroviral integrase containing
Length=1898
Score = 31.2 bits (69), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 34/150 (22%), Positives = 59/150 (39%), Gaps = 24/150 (16%)
Query 197 AGIGVFFGTDDRRYLFMPLVVGPQTYQRAELASILAALLRFVDYDDDSAKQQPSGRSEVR 256
AG G++ + + + P T A LA++ L RF G+S +
Sbjct 1325 AGFGLYVLSPTSPPVSLSFSCSPYTPTYAHLAAVACGLERF-------------GQSPLP 1371
Query 257 LVVISDSSYAVNCLGPWAARWCRNGWKSTSGKPVKNADLIQAILLLCDKRRKTAPSGATW 316
+V ++ ++ + L W G+ S+ G P+ + L+ I+ L SG +
Sbjct 1372 VVFLTHCNWIFSLLWELLPLWRARGFLSSDGAPLPHPSLLSYIISLT--------SGLS- 1422
Query 317 RGSVEYEYIRGHSGVYGNEMADKLAVQGAN 346
S+ + Y + G D LA QGA
Sbjct 1423 --SLPFIYRTSYRGSLFAVTVDTLAKQGAQ 1450
> cel:K12D12.5 hypothetical protein
Length=648
Score = 31.2 bits (69), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 22/56 (39%), Positives = 30/56 (53%), Gaps = 3/56 (5%)
Query 179 VEIYADGACPSNGRVDARAGIGVFFGTDDRRYLFMPLVVGPQTYQRAELASILAAL 234
V I+A C G+ DA A GV++G DD R LV QT RA + ++ +AL
Sbjct 166 VAIFA--ICEKEGKCDALAKYGVYWGQDDHRN-EAGLVEDGQTSLRAIMCAVRSAL 218
Lambda K H
0.318 0.134 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 16969450192
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40