bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0098_orf2
Length=224
Score E
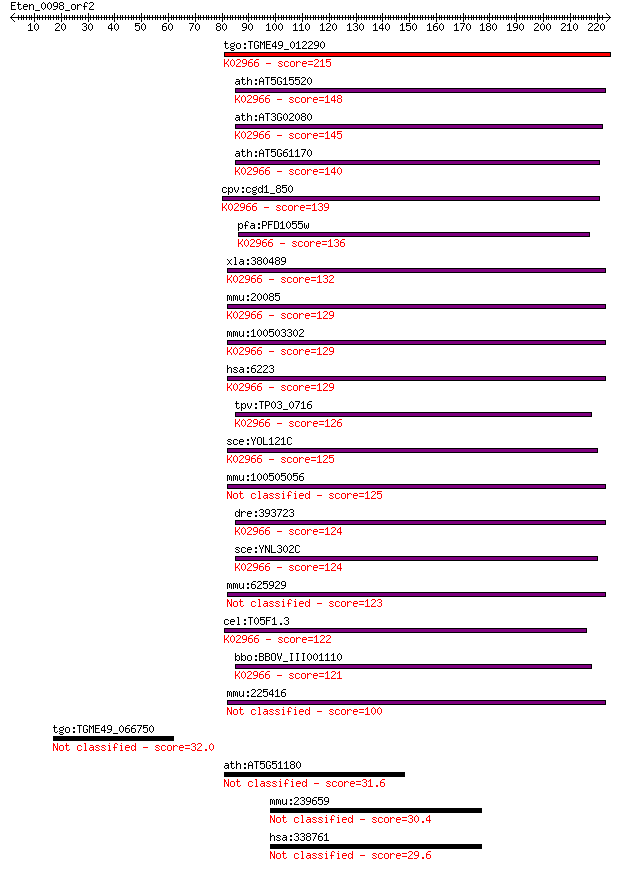
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_012290 40s ribosomal protein s19, putative ; K02966... 215 8e-56
ath:AT5G15520 40S ribosomal protein S19 (RPS19B); K02966 small... 148 2e-35
ath:AT3G02080 40S ribosomal protein S19 (RPS19A); K02966 small... 145 1e-34
ath:AT5G61170 40S ribosomal protein S19 (RPS19C); K02966 small... 140 3e-33
cpv:cgd1_850 40S ribosomal protein S19 ; K02966 small subunit ... 139 7e-33
pfa:PFD1055w 40S ribosomal protein S19, putative; K02966 small... 136 7e-32
xla:380489 rps19, MGC68643; ribosomal protein S19; K02966 smal... 132 1e-30
mmu:20085 Rps19, Dsk3, MGC107694; ribosomal protein S19; K0296... 129 1e-29
mmu:100503302 40S ribosomal protein S19 pseudogene; K02966 sma... 129 1e-29
hsa:6223 RPS19, DBA, DBA1; ribosomal protein S19; K02966 small... 129 1e-29
tpv:TP03_0716 40S ribosomal protein S19; K02966 small subunit ... 126 6e-29
sce:YOL121C RPS19A; Rps19ap; K02966 small subunit ribosomal pr... 125 1e-28
mmu:100505056 40S ribosomal protein S19-like 125 1e-28
dre:393723 rps19, MGC73211, zgc:73211; ribosomal protein S19; ... 124 2e-28
sce:YNL302C RPS19B, RP55B; Rps19bp; K02966 small subunit ribos... 124 2e-28
mmu:625929 Gm6636, EG625929; predicted gene 6636 123 6e-28
cel:T05F1.3 rps-19; Ribosomal Protein, Small subunit family me... 122 9e-28
bbo:BBOV_III001110 17.m07125; 40S ribosomal protein S19; K0296... 121 2e-27
mmu:225416 Gm4838, EG225416; predicted gene 4838 100 4e-21
tgo:TGME49_066750 hypothetical protein 32.0 1.7
ath:AT5G51180 hypothetical protein 31.6 2.5
mmu:239659 C1ql4, C1qtnf11; complement component 1, q subcompo... 30.4 5.7
hsa:338761 C1QL4, C1QTNF11, MGC131708; complement component 1,... 29.6 8.1
> tgo:TGME49_012290 40s ribosomal protein s19, putative ; K02966
small subunit ribosomal protein S19e
Length=160
Score = 215 bits (548), Expect = 8e-56, Method: Compositional matrix adjust.
Identities = 103/145 (71%), Positives = 116/145 (80%), Gaps = 1/145 (0%)
Query 81 TPVYTVKDVTAQTFIEAYANHLKKRGKFQVPMWADLVKTGYGKSLAPHDPDWIFKRAAAI 140
T V+TVKDV A FI YANHLKKRG+FQ+P W D VKT + K LAP+DPDW++ RAAAI
Sbjct 14 TRVHTVKDVKADAFIALYANHLKKRGRFQLPKWIDYVKTAHAKELAPYDPDWLYIRAAAI 73
Query 141 LRHLYLRPSCGVGAFRKVFSCKSGKKCSPGHTSKASGKIIRYILQQFETMGLVESRPDN- 199
LRHLY+RP CGVG FRKVFSC+ + P HTSKASGKIIRYILQQFE MGLVES P+
Sbjct 74 LRHLYIRPDCGVGGFRKVFSCRQRRGVRPNHTSKASGKIIRYILQQFEEMGLVESDPEKP 133
Query 200 GRRLTKVGQKELDVIARQCVAPSPA 224
GRRLTK GQ+ELDVIARQC P+ A
Sbjct 134 GRRLTKNGQRELDVIARQCAKPAVA 158
> ath:AT5G15520 40S ribosomal protein S19 (RPS19B); K02966 small
subunit ribosomal protein S19e
Length=143
Score = 148 bits (373), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 69/138 (50%), Positives = 94/138 (68%), Gaps = 0/138 (0%)
Query 85 TVKDVTAQTFIEAYANHLKKRGKFQVPMWADLVKTGYGKSLAPHDPDWIFKRAAAILRHL 144
TVKDV+ F++AYA+HLK+ GK ++P+W D+VKTG K LAP+DPDW + RAA++ R +
Sbjct 6 TVKDVSPHDFVKAYASHLKRSGKIELPLWTDIVKTGRLKELAPYDPDWYYIRAASMARKI 65
Query 145 YLRPSCGVGAFRKVFSCKSGKKCSPGHTSKASGKIIRYILQQFETMGLVESRPDNGRRLT 204
YLR GVGAFR+++ P H K+SG I R+ILQQ ETM +VE GRR+T
Sbjct 66 YLRGGLGVGAFRRIYGGSKRNGSRPPHFCKSSGGIARHILQQLETMSIVELDTKGGRRIT 125
Query 205 KVGQKELDVIARQCVAPS 222
GQ++LD +A + A S
Sbjct 126 SSGQRDLDQVAGRIAAES 143
> ath:AT3G02080 40S ribosomal protein S19 (RPS19A); K02966 small
subunit ribosomal protein S19e
Length=143
Score = 145 bits (365), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 70/138 (50%), Positives = 93/138 (67%), Gaps = 1/138 (0%)
Query 85 TVKDVTAQTFIEAYANHLKKRGKFQVPMWADLVKTGYGKSLAPHDPDWIFKRAAAILRHL 144
TVKDV+ F++AYA+HLK+ GK ++P W D+VKTG K LAP+DPDW + RAA++ R +
Sbjct 6 TVKDVSPHDFVKAYASHLKRSGKIELPTWTDIVKTGKLKELAPYDPDWYYIRAASMARKV 65
Query 145 YLRPSCGVGAFRKVFSCKSGKKCSPGHTSKASGKIIRYILQQFETMGLVESRPDNGRRLT 204
YLR GVGAFR+++ P H K+SG I R+ILQQ ETM +VE GRR+T
Sbjct 66 YLRGGLGVGAFRRIYGGSKRNGSRPPHFCKSSGGIARHILQQLETMNIVELDTKGGRRIT 125
Query 205 KVGQKELDVIA-RQCVAP 221
GQ++LD +A R V P
Sbjct 126 SSGQRDLDQVAGRIAVEP 143
> ath:AT5G61170 40S ribosomal protein S19 (RPS19C); K02966 small
subunit ribosomal protein S19e
Length=143
Score = 140 bits (354), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 64/136 (47%), Positives = 92/136 (67%), Gaps = 0/136 (0%)
Query 85 TVKDVTAQTFIEAYANHLKKRGKFQVPMWADLVKTGYGKSLAPHDPDWIFKRAAAILRHL 144
TVKDV+ F++AYA HLK+ GK ++P+W D+VKTG K LAP+DPDW + RAA++ R +
Sbjct 6 TVKDVSPHEFVKAYAAHLKRSGKIELPLWTDIVKTGKLKELAPYDPDWYYIRAASMARKV 65
Query 145 YLRPSCGVGAFRKVFSCKSGKKCSPGHTSKASGKIIRYILQQFETMGLVESRPDNGRRLT 204
YLR GVGAFR+++ P H K+SG + R+ILQQ +TM +V+ GR++T
Sbjct 66 YLRGGLGVGAFRRIYGGSKRNGSRPPHFCKSSGGVARHILQQLQTMNIVDLDTKGGRKIT 125
Query 205 KVGQKELDVIARQCVA 220
GQ++LD +A + A
Sbjct 126 SSGQRDLDQVAGRIAA 141
> cpv:cgd1_850 40S ribosomal protein S19 ; K02966 small subunit
ribosomal protein S19e
Length=154
Score = 139 bits (350), Expect = 7e-33, Method: Compositional matrix adjust.
Identities = 71/142 (50%), Positives = 90/142 (63%), Gaps = 1/142 (0%)
Query 80 PTPVYTVKDVTAQTFIEAYANHLKKRGKFQVPMWADLVKTGYGKSLAPHDPDWIFKRAAA 139
P + T KDV A IE +A LKK GK +P W D VKT K L P + DW++ RAA+
Sbjct 13 PRVIKTAKDVPAGILIEKFAQQLKKGGKMTMPEWVDHVKTASSKELPPQNSDWLYIRAAS 72
Query 140 ILRHLYLRPSCGVGAFRKVFSCKSGKKCSPGHTSKASGKIIRYILQQFETMGLVESRPDN 199
ILRHLY+RP+ GVGA K + K + H++ AS IIRY L Q +++GLVE P+
Sbjct 73 ILRHLYVRPNVGVGALSKFYGRKQRNGTARNHSAVASRGIIRYCLIQLQSLGLVEFNPNT 132
Query 200 G-RRLTKVGQKELDVIARQCVA 220
G RRL+ GQ+ELD IARQC A
Sbjct 133 GTRRLSPQGQRELDSIARQCKA 154
> pfa:PFD1055w 40S ribosomal protein S19, putative; K02966 small
subunit ribosomal protein S19e
Length=170
Score = 136 bits (342), Expect = 7e-32, Method: Compositional matrix adjust.
Identities = 68/132 (51%), Positives = 82/132 (62%), Gaps = 1/132 (0%)
Query 86 VKDVTAQTFIEAYANHLKKRGKFQVPMWADLVKTGYGKSLAPHDPDWIFKRAAAILRHLY 145
+KDV A FI +YA HLK K P W VKTG G+ LAP + DW F RA++ILR LY
Sbjct 29 IKDVDADLFIRSYATHLKLHNKITYPKWCTFVKTGKGRKLAPLNEDWYFIRASSILRRLY 88
Query 146 LRPSCGVGAFRKVFSCKSGKKCSPGHTSKASGKIIRYILQQFETMGLVESRP-DNGRRLT 204
L P GVG R+ FS K + +P HTS ASGKI+R ILQQ E +G VE P GRRLT
Sbjct 89 LHPDIGVGFLRRQFSSKQRRGVAPNHTSLASGKILRSILQQLENLGYVEQNPKKKGRRLT 148
Query 205 KVGQKELDVIAR 216
G+ ++ AR
Sbjct 149 TKGENAINNFAR 160
> xla:380489 rps19, MGC68643; ribosomal protein S19; K02966 small
subunit ribosomal protein S19e
Length=146
Score = 132 bits (331), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 63/141 (44%), Positives = 84/141 (59%), Gaps = 0/141 (0%)
Query 82 PVYTVKDVTAQTFIEAYANHLKKRGKFQVPMWADLVKTGYGKSLAPHDPDWIFKRAAAIL 141
P TVKDV Q F+ A A LKK GK +VP W D VK K LAP+D +W + RAA+ +
Sbjct 2 PGVTVKDVNQQEFVRALAAFLKKSGKLKVPEWVDTVKLAKHKELAPYDENWFYTRAASTV 61
Query 142 RHLYLRPSCGVGAFRKVFSCKSGKKCSPGHTSKASGKIIRYILQQFETMGLVESRPDNGR 201
RHLYLR GVG+ K++ + P H S+ S + R +LQ E++ +V+ P GR
Sbjct 62 RHLYLRGGAGVGSMTKIYGGRQSNGVMPSHFSRGSKSVARKVLQALESLKMVDKDPSGGR 121
Query 202 RLTKVGQKELDVIARQCVAPS 222
+LT GQ++LD IA Q A S
Sbjct 122 KLTPQGQRDLDRIAGQVAAAS 142
> mmu:20085 Rps19, Dsk3, MGC107694; ribosomal protein S19; K02966
small subunit ribosomal protein S19e
Length=145
Score = 129 bits (323), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 63/141 (44%), Positives = 82/141 (58%), Gaps = 0/141 (0%)
Query 82 PVYTVKDVTAQTFIEAYANHLKKRGKFQVPMWADLVKTGYGKSLAPHDPDWIFKRAAAIL 141
P TVKDV Q F+ A A LKK GK +VP W D VK K LAP+D +W + RAA+
Sbjct 2 PGVTVKDVNQQEFVRALAAFLKKSGKLKVPEWVDTVKLAKHKELAPYDENWFYTRAASTA 61
Query 142 RHLYLRPSCGVGAFRKVFSCKSGKKCSPGHTSKASGKIIRYILQQFETMGLVESRPDNGR 201
RHLYLR GVG+ K++ + P H S+ S + R +LQ E + +VE D GR
Sbjct 62 RHLYLRGGAGVGSMTKIYGGRQRNGVRPSHFSRGSKSVARRVLQALEGLKMVEKDQDGGR 121
Query 202 RLTKVGQKELDVIARQCVAPS 222
+LT GQ++LD IA Q A +
Sbjct 122 KLTPQGQRDLDRIAGQVAAAN 142
> mmu:100503302 40S ribosomal protein S19 pseudogene; K02966 small
subunit ribosomal protein S19e
Length=145
Score = 129 bits (323), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 63/141 (44%), Positives = 82/141 (58%), Gaps = 0/141 (0%)
Query 82 PVYTVKDVTAQTFIEAYANHLKKRGKFQVPMWADLVKTGYGKSLAPHDPDWIFKRAAAIL 141
P TVKDV Q F+ A A LKK GK +VP W D VK K LAP+D +W + RAA+
Sbjct 2 PGVTVKDVNQQEFVRALAAFLKKSGKLKVPEWVDTVKLAKHKELAPYDENWFYTRAASTA 61
Query 142 RHLYLRPSCGVGAFRKVFSCKSGKKCSPGHTSKASGKIIRYILQQFETMGLVESRPDNGR 201
RHLYLR GVG+ K++ + P H S+ S + R +LQ E + +VE D GR
Sbjct 62 RHLYLRGGAGVGSMTKIYGGRQRNGVRPSHFSRGSKSVARRVLQALEGLKMVEKDQDGGR 121
Query 202 RLTKVGQKELDVIARQCVAPS 222
+LT GQ++LD IA Q A +
Sbjct 122 KLTPQGQRDLDRIAGQVAAAN 142
> hsa:6223 RPS19, DBA, DBA1; ribosomal protein S19; K02966 small
subunit ribosomal protein S19e
Length=145
Score = 129 bits (323), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 63/141 (44%), Positives = 82/141 (58%), Gaps = 0/141 (0%)
Query 82 PVYTVKDVTAQTFIEAYANHLKKRGKFQVPMWADLVKTGYGKSLAPHDPDWIFKRAAAIL 141
P TVKDV Q F+ A A LKK GK +VP W D VK K LAP+D +W + RAA+
Sbjct 2 PGVTVKDVNQQEFVRALAAFLKKSGKLKVPEWVDTVKLAKHKELAPYDENWFYTRAASTA 61
Query 142 RHLYLRPSCGVGAFRKVFSCKSGKKCSPGHTSKASGKIIRYILQQFETMGLVESRPDNGR 201
RHLYLR GVG+ K++ + P H S+ S + R +LQ E + +VE D GR
Sbjct 62 RHLYLRGGAGVGSMTKIYGGRQRNGVMPSHFSRGSKSVARRVLQALEGLKMVEKDQDGGR 121
Query 202 RLTKVGQKELDVIARQCVAPS 222
+LT GQ++LD IA Q A +
Sbjct 122 KLTPQGQRDLDRIAGQVAAAN 142
> tpv:TP03_0716 40S ribosomal protein S19; K02966 small subunit
ribosomal protein S19e
Length=169
Score = 126 bits (317), Expect = 6e-29, Method: Compositional matrix adjust.
Identities = 60/134 (44%), Positives = 83/134 (61%), Gaps = 1/134 (0%)
Query 85 TVKDVTAQTFIEAYANHLKKRGKFQVPMWADLVKTGYGKSLAPHDPDWIFKRAAAILRHL 144
++KD F++A+A H+K +G + P D KT K LAP DPDW + RAAA+LRHL
Sbjct 27 SLKDCNPDLFVKAFAEHMKLKGYLECPKRLDYAKTSVAKELAPQDPDWFYIRAAAVLRHL 86
Query 145 YLRPSCGVGAFRKVFSCKSGKKCSPGHTSKASGKIIRYILQQFETMGLVESR-PDNGRRL 203
Y P GV RKV+S K +P HT +ASGKI+R I+QQ E++G +E +GRRL
Sbjct 87 YFHPDSGVDRLRKVYSSKKRNGSAPNHTCRASGKILRTIVQQLESIGFLEQDLKKHGRRL 146
Query 204 TKVGQKELDVIARQ 217
++ G ++ ARQ
Sbjct 147 SRKGCNTVNAFARQ 160
> sce:YOL121C RPS19A; Rps19ap; K02966 small subunit ribosomal
protein S19e
Length=144
Score = 125 bits (314), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 59/139 (42%), Positives = 87/139 (62%), Gaps = 1/139 (0%)
Query 82 PVYTVKDVTAQTFIEAYANHLKKRGKFQVPMWADLVKTGYGKSLAPHDPD-WIFKRAAAI 140
P +V+DV AQ FI AYA+ L+++GK +VP + D+VKT G + P D + W +KRAA++
Sbjct 2 PGVSVRDVAAQDFINAYASFLQRQGKLEVPGYVDIVKTSSGNEMPPQDAEGWFYKRAASV 61
Query 141 LRHLYLRPSCGVGAFRKVFSCKSGKKCSPGHTSKASGKIIRYILQQFETMGLVESRPDNG 200
RH+Y+R GVG K++ + P ASG I R +LQ E +G+VE P G
Sbjct 62 ARHIYMRKQVGVGKLNKLYGGAKSRGVRPYKHIDASGSINRKVLQALEKIGIVEISPKGG 121
Query 201 RRLTKVGQKELDVIARQCV 219
RR+++ GQ++LD IA Q +
Sbjct 122 RRISENGQRDLDRIAAQTL 140
> mmu:100505056 40S ribosomal protein S19-like
Length=145
Score = 125 bits (313), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 62/141 (43%), Positives = 81/141 (57%), Gaps = 0/141 (0%)
Query 82 PVYTVKDVTAQTFIEAYANHLKKRGKFQVPMWADLVKTGYGKSLAPHDPDWIFKRAAAIL 141
P TVKDV Q F+ A A LKK K +VP W D VK K LAP+D +W + RAA+
Sbjct 2 PGVTVKDVNQQEFVRALAAFLKKSRKLKVPEWVDTVKLAKHKELAPYDENWFYTRAASTA 61
Query 142 RHLYLRPSCGVGAFRKVFSCKSGKKCSPGHTSKASGKIIRYILQQFETMGLVESRPDNGR 201
HLYLR GVG+ K++ + P H S+ S + R +LQ E + +VE D GR
Sbjct 62 PHLYLRGGAGVGSMTKIYGGRQRNGVRPSHFSRGSKSVARRVLQALEGLKMVEKDQDGGR 121
Query 202 RLTKVGQKELDVIARQCVAPS 222
+LT GQ++LD IARQ A +
Sbjct 122 KLTPQGQRDLDRIARQVAAAN 142
> dre:393723 rps19, MGC73211, zgc:73211; ribosomal protein S19;
K02966 small subunit ribosomal protein S19e
Length=146
Score = 124 bits (312), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 63/138 (45%), Positives = 81/138 (58%), Gaps = 0/138 (0%)
Query 85 TVKDVTAQTFIEAYANHLKKRGKFQVPMWADLVKTGYGKSLAPHDPDWIFKRAAAILRHL 144
TVKDV Q F+ A A LKK GK +VP W D+VK K LAP D +W + RAA+ +RHL
Sbjct 6 TVKDVNQQEFVRALAAFLKKSGKLKVPDWVDIVKLAKHKELAPCDENWFYIRAASTVRHL 65
Query 145 YLRPSCGVGAFRKVFSCKSGKKCSPGHTSKASGKIIRYILQQFETMGLVESRPDNGRRLT 204
YLR GVG+ K++ + P H S S + R +LQ E + +VE P+ GRRLT
Sbjct 66 YLRGGVGVGSMTKIYGGRKRNGVCPSHFSVGSKNVARKVLQALEGLKMVEKDPNGGRRLT 125
Query 205 KVGQKELDVIARQCVAPS 222
G ++LD IA Q A S
Sbjct 126 PQGTRDLDRIAGQVAAAS 143
> sce:YNL302C RPS19B, RP55B; Rps19bp; K02966 small subunit ribosomal
protein S19e
Length=144
Score = 124 bits (312), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 58/136 (42%), Positives = 86/136 (63%), Gaps = 1/136 (0%)
Query 85 TVKDVTAQTFIEAYANHLKKRGKFQVPMWADLVKTGYGKSLAPHDPD-WIFKRAAAILRH 143
+V+DV AQ FI AYA+ L+++GK +VP + D+VKT G + P D + W +KRAA++ RH
Sbjct 5 SVRDVAAQDFINAYASFLQRQGKLEVPGYVDIVKTSSGNEMPPQDAEGWFYKRAASVARH 64
Query 144 LYLRPSCGVGAFRKVFSCKSGKKCSPGHTSKASGKIIRYILQQFETMGLVESRPDNGRRL 203
+Y+R GVG K++ + P ASG I R +LQ E +G+VE P GRR+
Sbjct 65 IYMRKQVGVGKLNKLYGGAKSRGVRPYKHIDASGSINRKVLQALEKIGIVEISPKGGRRI 124
Query 204 TKVGQKELDVIARQCV 219
++ GQ++LD IA Q +
Sbjct 125 SENGQRDLDRIAAQTL 140
> mmu:625929 Gm6636, EG625929; predicted gene 6636
Length=145
Score = 123 bits (308), Expect = 6e-28, Method: Compositional matrix adjust.
Identities = 61/141 (43%), Positives = 80/141 (56%), Gaps = 0/141 (0%)
Query 82 PVYTVKDVTAQTFIEAYANHLKKRGKFQVPMWADLVKTGYGKSLAPHDPDWIFKRAAAIL 141
P TVKDV Q F A A LKK GK +VP W D VK K LAP+D +W + RAA+
Sbjct 2 PGVTVKDVNQQEFTRALAAFLKKSGKLKVPEWVDTVKLAKHKELAPYDENWFYTRAASTA 61
Query 142 RHLYLRPSCGVGAFRKVFSCKSGKKCSPGHTSKASGKIIRYILQQFETMGLVESRPDNGR 201
H+YLR GVG+ K++ + P H S+ S + R +LQ E + +VE D GR
Sbjct 62 WHVYLRGGAGVGSMTKIYGGRQRNGVRPSHFSRGSKSVARQVLQALEGLKMVEKDQDGGR 121
Query 202 RLTKVGQKELDVIARQCVAPS 222
+LT GQ++LD IA Q A +
Sbjct 122 KLTPQGQRDLDRIAGQVAAAN 142
> cel:T05F1.3 rps-19; Ribosomal Protein, Small subunit family
member (rps-19); K02966 small subunit ribosomal protein S19e
Length=146
Score = 122 bits (307), Expect = 9e-28, Method: Compositional matrix adjust.
Identities = 62/136 (45%), Positives = 85/136 (62%), Gaps = 2/136 (1%)
Query 81 TPVYTVKDVTAQTFIEAYANHLKKRGKFQVPMWADLVKTGYGKSLAPHDPDWIFKRAAAI 140
T ++KDV ++ A+ LKK GK +VP W+DLVK G K LAP DPDW + RAA++
Sbjct 2 TRATSIKDVDQHEATKSIAHFLKKSGKVKVPEWSDLVKLGVNKELAPVDPDWFYTRAASL 61
Query 141 LRHLYLRPSCGVGAFRKVFSCKSGKKCSPGHTSKASGKIIRYILQQFETMGLVESRPD-N 199
RHLY RP G+GAF+KV+ + +P H ++G +R +QQ E + VE PD
Sbjct 62 ARHLYFRP-AGIGAFKKVYGGNKRRGVAPNHFQTSAGNCLRKAVQQLEKIKWVEKHPDGK 120
Query 200 GRRLTKVGQKELDVIA 215
GR L+K G+K+LD IA
Sbjct 121 GRILSKQGRKDLDRIA 136
> bbo:BBOV_III001110 17.m07125; 40S ribosomal protein S19; K02966
small subunit ribosomal protein S19e
Length=171
Score = 121 bits (303), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 58/134 (43%), Positives = 79/134 (58%), Gaps = 1/134 (0%)
Query 85 TVKDVTAQTFIEAYANHLKKRGKFQVPMWADLVKTGYGKSLAPHDPDWIFKRAAAILRHL 144
++ D FI A+A ++ + P W D VKT K LAP DP W + R+AAILRHL
Sbjct 27 SLNDCQPDVFIAAFAQQMELKNYIDPPKWIDYVKTSCAKELAPQDPKWFYIRSAAILRHL 86
Query 145 YLRPSCGVGAFRKVFSCKSGKKCSPGHTSKASGKIIRYILQQFETMGLVESRPDN-GRRL 203
Y P G+G R+V++ + + C+P HT ASGKIIR I+QQ E G +E DN GRRL
Sbjct 87 YFHPDEGIGRLRRVYASRKRRGCAPNHTCPASGKIIRTIVQQLEANGFLEKSKDNKGRRL 146
Query 204 TKVGQKELDVIARQ 217
++ G ++ AR
Sbjct 147 SRRGCNIVNEFARH 160
> mmu:225416 Gm4838, EG225416; predicted gene 4838
Length=145
Score = 100 bits (249), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 55/141 (39%), Positives = 72/141 (51%), Gaps = 0/141 (0%)
Query 82 PVYTVKDVTAQTFIEAYANHLKKRGKFQVPMWADLVKTGYGKSLAPHDPDWIFKRAAAIL 141
P TVKDV Q + A A KK K V W + VK K LAP+D W + RAA+
Sbjct 2 PRVTVKDVNQQEVVRALAASFKKSRKLIVSKWVNTVKLTKHKELAPYDEKWFYTRAASTA 61
Query 142 RHLYLRPSCGVGAFRKVFSCKSGKKCSPGHTSKASGKIIRYILQQFETMGLVESRPDNGR 201
RHLYLR GVG+ K++ + P S+ S + R +LQ E + + + D G
Sbjct 62 RHLYLRGCAGVGSMTKIYRGPQRNRNRPRLFSRGSKSVARRVLQALEGLKMEDQDQDGGH 121
Query 202 RLTKVGQKELDVIARQCVAPS 222
LT GQ++LD IA Q A S
Sbjct 122 NLTPQGQRDLDKIAGQVAAVS 142
> tgo:TGME49_066750 hypothetical protein
Length=355
Score = 32.0 bits (71), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 17/47 (36%), Positives = 25/47 (53%), Gaps = 2/47 (4%)
Query 17 VRAVFIREHITSVGGAIIFKTAYGASALQNDAQEVN--CKPARFILL 61
+RAV R+H VG ++F AY A L D ++N P+ F +L
Sbjct 9 LRAVLTRQHAIEVGVVLLFLVAYSAQPLLVDVIKINGGAHPSTFSVL 55
> ath:AT5G51180 hypothetical protein
Length=357
Score = 31.6 bits (70), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 27/67 (40%), Gaps = 6/67 (8%)
Query 81 TPVYTVKDVTAQTFIEAYANHLKKRGKFQVPMWADLVKTGYGKSLAPHDPDWIFKRAAAI 140
TP T+ + A FI HL G QVP L H WIFKR
Sbjct 145 TPKGTICGLEAMNFITVATPHLGSMGNKQVPFLFGFSSIEKVAGLIIH---WIFKRTG-- 199
Query 141 LRHLYLR 147
RHL+L+
Sbjct 200 -RHLFLK 205
> mmu:239659 C1ql4, C1qtnf11; complement component 1, q subcomponent-like
4
Length=238
Score = 30.4 bits (67), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 27/81 (33%), Positives = 40/81 (49%), Gaps = 10/81 (12%)
Query 98 YANHLKKRGKFQVPMWADLVKTGYGKSLA-PHDPDWIFKRAA-AILRHLYLRPSCGVGAF 155
+A H+ RG MWADL+K G ++ A D D + A+ +++ HL VG
Sbjct 156 FAYHVLMRGGDGTSMWADLMKNGQVRASAIAQDADQNYDYASNSVILHL------DVG-- 207
Query 156 RKVFSCKSGKKCSPGHTSKAS 176
+VF G K G+T+K S
Sbjct 208 DEVFIKLDGGKVHGGNTNKYS 228
> hsa:338761 C1QL4, C1QTNF11, MGC131708; complement component
1, q subcomponent-like 4
Length=238
Score = 29.6 bits (65), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 39/81 (48%), Gaps = 10/81 (12%)
Query 98 YANHLKKRGKFQVPMWADLVKTGYGKSLA-PHDPDWIFKRAA-AILRHLYLRPSCGVGAF 155
+A H+ RG MWADL+K G ++ A D D + A+ +++ HL +
Sbjct 156 FAYHVLMRGGDGTSMWADLMKNGQVRASAIAQDADQNYDYASNSVILHLDVG-------- 207
Query 156 RKVFSCKSGKKCSPGHTSKAS 176
+VF G K G+T+K S
Sbjct 208 DEVFIKLDGGKVHGGNTNKYS 228
Lambda K H
0.323 0.138 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7459475120
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40