bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0106_orf2
Length=271
Score E
Sequences producing significant alignments: (Bits) Value
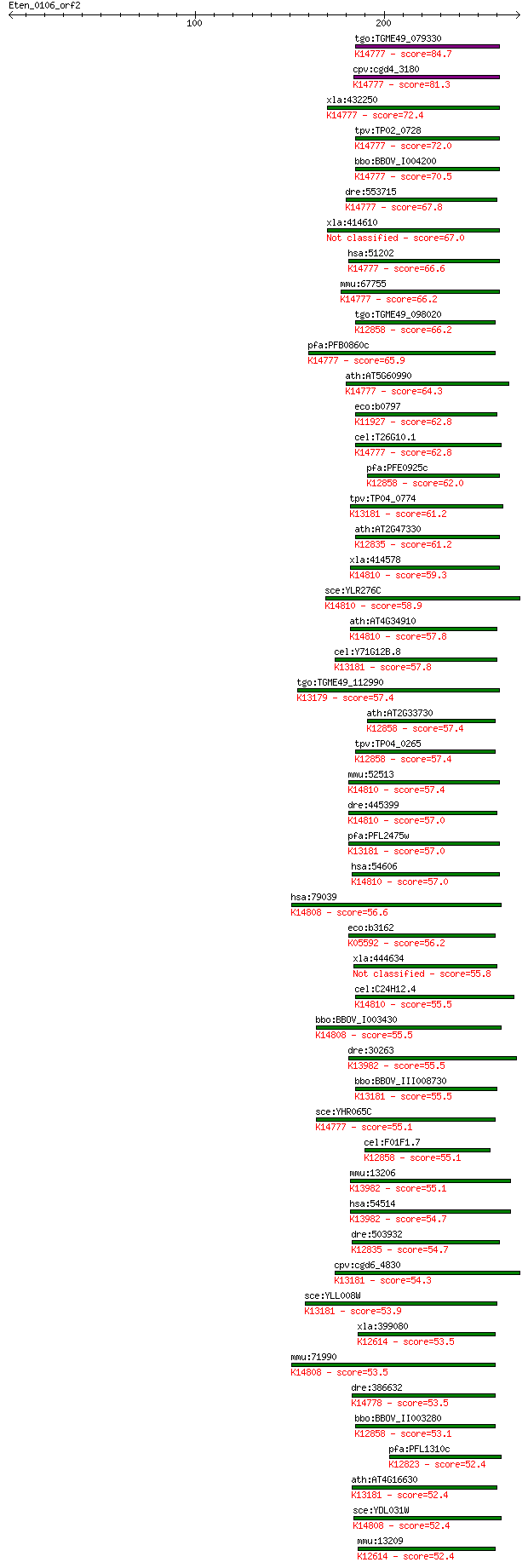
tgo:TGME49_079330 ATP-dependent RNA helicase, putative ; K1477... 84.7 3e-16
cpv:cgd4_3180 Rrp3p, eIF4A-1-family RNA SFII helicase (DEXDc+H... 81.3 4e-15
xla:432250 ddx47, MGC81303; DEAD (Asp-Glu-Ala-Asp) box polypep... 72.4 2e-12
tpv:TP02_0728 ATP-dependent RNA helicase; K14777 ATP-dependent... 72.0 2e-12
bbo:BBOV_I004200 19.m02020; DEAD/DEAH box helicase and helicas... 70.5 6e-12
dre:553715 MGC112350; zgc:112350 (EC:3.6.4.13); K14777 ATP-dep... 67.8 5e-11
xla:414610 hypothetical protein MGC81500 67.0 6e-11
hsa:51202 DDX47, DKFZp564O176, E4-DBP, FLJ30012, HQ0256, MSTP1... 66.6 9e-11
mmu:67755 Ddx47, 4930588A18Rik, C77285; DEAD (Asp-Glu-Ala-Asp)... 66.2 1e-10
tgo:TGME49_098020 DEAD-box ATP-dependent RNA helicase, putativ... 66.2 1e-10
pfa:PFB0860c DEAD/DEAH box helicase, putative; K14777 ATP-depe... 65.9 2e-10
ath:AT5G60990 DEAD/DEAH box helicase, putative (RH10); K14777 ... 64.3 4e-10
eco:b0797 rhlE, ECK0786, JW0781; ATP-dependent RNA helicase; K... 62.8 1e-09
cel:T26G10.1 hypothetical protein; K14777 ATP-dependent RNA he... 62.8 1e-09
pfa:PFE0925c snrnp protein, putative; K12858 ATP-dependent RNA... 62.0 2e-09
tpv:TP04_0774 DEAD box RNA helicase (EC:3.6.1.-); K13181 ATP-d... 61.2 3e-09
ath:AT2G47330 DEAD/DEAH box helicase, putative; K12835 ATP-dep... 61.2 3e-09
xla:414578 ddx56, MGC81243; DEAD (Asp-Glu-Ala-Asp) box polypep... 59.3 1e-08
sce:YLR276C DBP9; ATP-dependent RNA helicase of the DEAD-box f... 58.9 2e-08
ath:AT4G34910 DEAD/DEAH box helicase, putative (RH16); K14810 ... 57.8 4e-08
cel:Y71G12B.8 hypothetical protein; K13181 ATP-dependent RNA h... 57.8 4e-08
tgo:TGME49_112990 ATP-dependent RNA helicase, putative (EC:5.9... 57.4 5e-08
ath:AT2G33730 DEAD box RNA helicase, putative; K12858 ATP-depe... 57.4 5e-08
tpv:TP04_0265 small nuclear ribonucleoprotein; K12858 ATP-depe... 57.4 6e-08
mmu:52513 Ddx56, 2600001H07Rik, D11Ertd619e, Noh61; DEAD (Asp-... 57.4 6e-08
dre:445399 ddx56, noh61, zgc:110078; DEAD (Asp-Glu-Ala-Asp) bo... 57.0 7e-08
pfa:PFL2475w DEAD/DEAH box helicase, putative; K13181 ATP-depe... 57.0 7e-08
hsa:54606 DDX56, DDX21, DDX26, NOH61; DEAD (Asp-Glu-Ala-Asp) b... 57.0 8e-08
hsa:79039 DDX54, DP97, MGC2835; DEAD (Asp-Glu-Ala-Asp) box pol... 56.6 9e-08
eco:b3162 deaD, csdA, ECK3150, JW5531, mssB, rhlD; ATP-depende... 56.2 1e-07
xla:444634 MGC84147 protein 55.8 2e-07
cel:C24H12.4 hypothetical protein; K14810 ATP-dependent RNA he... 55.5 2e-07
bbo:BBOV_I003430 19.m02227; DEAD/DEAH box helicase and helicas... 55.5 2e-07
dre:30263 vasa, MGC158535, fi24g05, vas, vlg, wu:fi24g05, zgc:... 55.5 2e-07
bbo:BBOV_III008730 17.m07763; DEAD/DEAH box helicase domain co... 55.5 2e-07
sce:YHR065C RRP3; Protein involved in rRNA processing; require... 55.1 2e-07
cel:F01F1.7 ddx-23; DEAD boX helicase homolog family member (d... 55.1 3e-07
mmu:13206 Ddx4, AV206478, Mvh, VASA; DEAD (Asp-Glu-Ala-Asp) bo... 55.1 3e-07
hsa:54514 DDX4, MGC111074, VASA; DEAD (Asp-Glu-Ala-Asp) box po... 54.7 4e-07
dre:503932 ddx42, im:7148194, zgc:111815; DEAD (Asp-Glu-Ala-As... 54.7 4e-07
cpv:cgd6_4830 Drs1p, eIF4a-1-family RNA SFII helicase ; K13181... 54.3 4e-07
sce:YLL008W DRS1; Drs1p (EC:3.6.1.-); K13181 ATP-dependent RNA... 53.9 5e-07
xla:399080 ddx6, p54h; DEAD (Asp-Glu-Ala-Asp) box polypeptide ... 53.5 9e-07
mmu:71990 Ddx54, 2410015A15Rik, AI414901, DP97; DEAD (Asp-Glu-... 53.5 9e-07
dre:386632 ddx49, wu:fb82g04, wu:fd12e05; DEAD (Asp-Glu-Ala-As... 53.5 9e-07
bbo:BBOV_II003280 18.m06276; DEAD box RNA helicase; K12858 ATP... 53.1 1e-06
pfa:PFL1310c DEAD-box helicase; K12823 ATP-dependent RNA helic... 52.4 2e-06
ath:AT4G16630 DEAD/DEAH box helicase, putative (RH28); K13181 ... 52.4 2e-06
sce:YDL031W DBP10; Dbp10p (EC:3.6.1.-); K14808 ATP-dependent R... 52.4 2e-06
mmu:13209 Ddx6, 1110001P04Rik, E230023J21Rik, HLR2, mRCK/P54, ... 52.4 2e-06
> tgo:TGME49_079330 ATP-dependent RNA helicase, putative ; K14777
ATP-dependent RNA helicase DDX47/RRP3 [EC:3.6.4.13]
Length=479
Score = 84.7 bits (208), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 41/76 (53%), Positives = 51/76 (67%), Gaps = 12/76 (15%)
Query 185 TFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAATGS 244
TFASLGLC E+CA+ + W+ PTA+Q +V+P ALQ RD+IA+A TGS
Sbjct 52 TFASLGLCSELCASVSTLGWKSPTAIQSEVLPYALQ------------GRDIIALAETGS 99
Query 245 GKTXAFVLPLLQHLLQ 260
GKT AF LP+LQ LLQ
Sbjct 100 GKTAAFGLPILQRLLQ 115
> cpv:cgd4_3180 Rrp3p, eIF4A-1-family RNA SFII helicase (DEXDc+HELICc)
; K14777 ATP-dependent RNA helicase DDX47/RRP3 [EC:3.6.4.13]
Length=446
Score = 81.3 bits (199), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 35/77 (45%), Positives = 50/77 (64%), Gaps = 12/77 (15%)
Query 184 ATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAATG 243
TFASLG+C+E+C A + W+ PT +Q++ IP+AL E RD+I +A TG
Sbjct 33 VTFASLGVCKELCIACESLGWKTPTEIQKKTIPVAL------------EGRDIIGLAETG 80
Query 244 SGKTXAFVLPLLQHLLQ 260
SGKT +F++P+LQ LL
Sbjct 81 SGKTGSFIIPILQRLLD 97
> xla:432250 ddx47, MGC81303; DEAD (Asp-Glu-Ala-Asp) box polypeptide
47; K14777 ATP-dependent RNA helicase DDX47/RRP3 [EC:3.6.4.13]
Length=448
Score = 72.4 bits (176), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 36/91 (39%), Positives = 50/91 (54%), Gaps = 12/91 (13%)
Query 170 QQQQDGAAAGGEAAATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVR 229
+++ D G E TF LG+ +C A W++PT +Q + IP+ALQ
Sbjct 5 EEEHDVVENGEEEPKTFRDLGVTDVLCEACEQLGWKQPTKIQIEAIPMALQ--------- 55
Query 230 TVEQRDVIAVAATGSGKTXAFVLPLLQHLLQ 260
RD+I +A TGSGKT AF LP+LQ LL+
Sbjct 56 ---GRDIIGLAETGSGKTGAFALPILQTLLE 83
> tpv:TP02_0728 ATP-dependent RNA helicase; K14777 ATP-dependent
RNA helicase DDX47/RRP3 [EC:3.6.4.13]
Length=470
Score = 72.0 bits (175), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 33/76 (43%), Positives = 45/76 (59%), Gaps = 12/76 (15%)
Query 185 TFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAATGS 244
TF LG+C E+C A W+RPT +Q + IP+AL +D+I +A TGS
Sbjct 42 TFEDLGVCVELCRACKELGWKRPTKIQIEAIPIALS------------GKDIIGLAETGS 89
Query 245 GKTXAFVLPLLQHLLQ 260
GKT AF +P+LQ LL+
Sbjct 90 GKTAAFTIPILQKLLE 105
> bbo:BBOV_I004200 19.m02020; DEAD/DEAH box helicase and helicase
conserved C-terminal domain containing protein (EC:3.6.1.-);
K14777 ATP-dependent RNA helicase DDX47/RRP3 [EC:3.6.4.13]
Length=433
Score = 70.5 bits (171), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 35/76 (46%), Positives = 44/76 (57%), Gaps = 12/76 (15%)
Query 185 TFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAATGS 244
TF SLG+C E+C A + W+ PT +Q IP AL RDVI +A TGS
Sbjct 44 TFQSLGVCPELCKACQSMGWQAPTPIQMAAIPHALN------------GRDVIGLAVTGS 91
Query 245 GKTXAFVLPLLQHLLQ 260
GKT AF +P+L HLL+
Sbjct 92 GKTGAFTIPVLHHLLE 107
> dre:553715 MGC112350; zgc:112350 (EC:3.6.4.13); K14777 ATP-dependent
RNA helicase DDX47/RRP3 [EC:3.6.4.13]
Length=512
Score = 67.8 bits (164), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 34/80 (42%), Positives = 45/80 (56%), Gaps = 12/80 (15%)
Query 180 GEAAATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAV 239
GE +F LG+ +C A W++PT +Q + IP+ALQ RDVI +
Sbjct 75 GEIHTSFKELGVTEVLCEACDQLGWKKPTKIQIEAIPVALQG------------RDVIGL 122
Query 240 AATGSGKTXAFVLPLLQHLL 259
A TGSGKT AF +P+LQ LL
Sbjct 123 AETGSGKTGAFAVPVLQSLL 142
> xla:414610 hypothetical protein MGC81500
Length=317
Score = 67.0 bits (162), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 35/91 (38%), Positives = 48/91 (52%), Gaps = 12/91 (13%)
Query 170 QQQQDGAAAGGEAAATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVR 229
+ + D E TF LG+ +C A W++PT +Q + IP+ALQ
Sbjct 5 EDEHDVLENAEEEQKTFRDLGVTDVLCEACEQLGWKQPTKIQIEAIPMALQG-------- 56
Query 230 TVEQRDVIAVAATGSGKTXAFVLPLLQHLLQ 260
RD+I +A TGSGKT AF LP+LQ LL+
Sbjct 57 ----RDIIGLAETGSGKTGAFALPILQTLLE 83
> hsa:51202 DDX47, DKFZp564O176, E4-DBP, FLJ30012, HQ0256, MSTP162,
RRP3; DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 (EC:3.6.4.13);
K14777 ATP-dependent RNA helicase DDX47/RRP3 [EC:3.6.4.13]
Length=455
Score = 66.6 bits (161), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 34/80 (42%), Positives = 43/80 (53%), Gaps = 12/80 (15%)
Query 181 EAAATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVA 240
E TF LG+ +C A W +PT +Q + IPLALQ RD+I +A
Sbjct 21 EETKTFKDLGVTDVLCEACDQLGWTKPTKIQIEAIPLALQG------------RDIIGLA 68
Query 241 ATGSGKTXAFVLPLLQHLLQ 260
TGSGKT AF LP+L LL+
Sbjct 69 ETGSGKTGAFALPILNALLE 88
> mmu:67755 Ddx47, 4930588A18Rik, C77285; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 47 (EC:3.6.4.13); K14777 ATP-dependent RNA
helicase DDX47/RRP3 [EC:3.6.4.13]
Length=455
Score = 66.2 bits (160), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 36/84 (42%), Positives = 45/84 (53%), Gaps = 12/84 (14%)
Query 177 AAGGEAAATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDV 236
AA E TF LG+ +C A W +PT +Q + IPLALQ RD+
Sbjct 17 AAEEEETKTFKDLGVTDVLCEACDQLGWAKPTKIQIEAIPLALQ------------GRDI 64
Query 237 IAVAATGSGKTXAFVLPLLQHLLQ 260
I +A TGSGKT AF LP+L LL+
Sbjct 65 IGLAETGSGKTGAFALPILNALLE 88
> tgo:TGME49_098020 DEAD-box ATP-dependent RNA helicase, putative
(EC:2.7.11.25); K12858 ATP-dependent RNA helicase DDX23/PRP28
[EC:3.6.4.13]
Length=1158
Score = 66.2 bits (160), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 33/74 (44%), Positives = 44/74 (59%), Gaps = 12/74 (16%)
Query 185 TFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAATGS 244
T+A L E+ A A ++RPT +Q Q IP+AL EQRD+I +A TGS
Sbjct 738 TWAESALPWELIEAVKHANYDRPTPIQMQAIPIAL------------EQRDLIGIAETGS 785
Query 245 GKTXAFVLPLLQHL 258
GKT AFVLP+L ++
Sbjct 786 GKTAAFVLPMLTYV 799
> pfa:PFB0860c DEAD/DEAH box helicase, putative; K14777 ATP-dependent
RNA helicase DDX47/RRP3 [EC:3.6.4.13]
Length=562
Score = 65.9 bits (159), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 27/99 (27%), Positives = 52/99 (52%), Gaps = 21/99 (21%)
Query 160 EAQGVEQQQEQQQQDGAAAGGEAAATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLAL 219
E + + +E+++Q+ TF L +C E+ + W++PT +Q++++P A
Sbjct 141 EVKNLVTNEEREKQN---------VTFEDLNICEEILESIKELGWKKPTEIQREILPHAF 191
Query 220 QQLQRLRGVRTVEQRDVIAVAATGSGKTXAFVLPLLQHL 258
+ +D+I ++ TGSGKT F++P+LQ L
Sbjct 192 LK------------KDIIGLSETGSGKTACFIIPILQDL 218
> ath:AT5G60990 DEAD/DEAH box helicase, putative (RH10); K14777
ATP-dependent RNA helicase DDX47/RRP3 [EC:3.6.4.13]
Length=456
Score = 64.3 bits (155), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 33/86 (38%), Positives = 46/86 (53%), Gaps = 12/86 (13%)
Query 180 GEAAATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAV 239
E TFA LG+ E+ A W+ P+ +Q + +P AL E +DVI +
Sbjct 5 NEVVKTFAELGVREELVKACERLGWKNPSKIQAEALPFAL------------EGKDVIGL 52
Query 240 AATGSGKTXAFVLPLLQHLLQHGSSS 265
A TGSGKT AF +P+LQ LL++ S
Sbjct 53 AQTGSGKTGAFAIPILQALLEYVYDS 78
> eco:b0797 rhlE, ECK0786, JW0781; ATP-dependent RNA helicase;
K11927 ATP-dependent RNA helicase RhlE [EC:3.6.4.13]
Length=454
Score = 62.8 bits (151), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 33/75 (44%), Positives = 42/75 (56%), Gaps = 12/75 (16%)
Query 185 TFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAATGS 244
+F SLGL ++ A A + PT +QQQ IP L E RD++A A TG+
Sbjct 2 SFDSLGLSPDILRAVAEQGYREPTPIQQQAIPAVL------------EGRDLMASAQTGT 49
Query 245 GKTXAFVLPLLQHLL 259
GKT F LPLLQHL+
Sbjct 50 GKTAGFTLPLLQHLI 64
> cel:T26G10.1 hypothetical protein; K14777 ATP-dependent RNA
helicase DDX47/RRP3 [EC:3.6.4.13]
Length=489
Score = 62.8 bits (151), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 32/77 (41%), Positives = 44/77 (57%), Gaps = 12/77 (15%)
Query 185 TFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAATGS 244
+FA LG+ + +C A W +P+ +QQ +P ALQ +DVI +A TGS
Sbjct 45 SFAELGVSQPLCDACQRLGWMKPSKIQQAALPHALQG------------KDVIGLAETGS 92
Query 245 GKTXAFVLPLLQHLLQH 261
GKT AF +P+LQ LL H
Sbjct 93 GKTGAFAIPVLQSLLDH 109
> pfa:PFE0925c snrnp protein, putative; K12858 ATP-dependent RNA
helicase DDX23/PRP28 [EC:3.6.4.13]
Length=1123
Score = 62.0 bits (149), Expect = 2e-09, Method: Composition-based stats.
Identities = 31/70 (44%), Positives = 42/70 (60%), Gaps = 12/70 (17%)
Query 191 LCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAATGSGKTXAF 250
L ++ A AK+E+PT +Q Q IP+AL E RD+I +A TGSGKT AF
Sbjct 705 LSNDLLKAIKKAKYEKPTPIQMQAIPIAL------------EMRDLIGIAETGSGKTAAF 752
Query 251 VLPLLQHLLQ 260
VLP+L ++ Q
Sbjct 753 VLPMLSYVKQ 762
> tpv:TP04_0774 DEAD box RNA helicase (EC:3.6.1.-); K13181 ATP-dependent
RNA helicase DDX27 [EC:3.6.4.13]
Length=543
Score = 61.2 bits (147), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 45/81 (55%), Gaps = 12/81 (14%)
Query 182 AAATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAA 241
+ ++ GLCR + A + ++ PT +Q +VIPLAL E +D++ A
Sbjct 73 SNLNWSDFGLCRSILRAISEMGYQNPTIIQSKVIPLAL------------EGKDLLVTAE 120
Query 242 TGSGKTXAFVLPLLQHLLQHG 262
TGSGKT +F++P LQ L+ G
Sbjct 121 TGSGKTASFLIPTLQRLVVSG 141
> ath:AT2G47330 DEAD/DEAH box helicase, putative; K12835 ATP-dependent
RNA helicase DDX42 [EC:3.6.4.13]
Length=760
Score = 61.2 bits (147), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 29/76 (38%), Positives = 42/76 (55%), Gaps = 12/76 (15%)
Query 185 TFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAATGS 244
TF G ++ +A +E+PTA+Q Q +P+ L RDVI +A TGS
Sbjct 229 TFEDCGFSSQIMSAIKKQAYEKPTAIQCQALPIVLSG------------RDVIGIAKTGS 276
Query 245 GKTXAFVLPLLQHLLQ 260
GKT AFVLP++ H++
Sbjct 277 GKTAAFVLPMIVHIMD 292
> xla:414578 ddx56, MGC81243; DEAD (Asp-Glu-Ala-Asp) box polypeptide
56; K14810 ATP-dependent RNA helicase DDX56/DBP9 [EC:3.6.4.13]
Length=552
Score = 59.3 bits (142), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 31/79 (39%), Positives = 46/79 (58%), Gaps = 12/79 (15%)
Query 182 AAATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAA 241
AA F LGL + + A W +PT +Q++ IPLAL E +D++A A
Sbjct 2 AALQFHELGLDDRLLKSIADLGWAKPTLIQEKAIPLAL------------EGKDLLARAR 49
Query 242 TGSGKTXAFVLPLLQHLLQ 260
TGSGKT ++ +P++Q+LLQ
Sbjct 50 TGSGKTASYSIPIIQNLLQ 68
> sce:YLR276C DBP9; ATP-dependent RNA helicase of the DEAD-box
family involved in biogenesis of the 60S ribosomal subunit
(EC:3.6.1.-); K14810 ATP-dependent RNA helicase DDX56/DBP9 [EC:3.6.4.13]
Length=594
Score = 58.9 bits (141), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 32/103 (31%), Positives = 56/103 (54%), Gaps = 13/103 (12%)
Query 169 EQQQQDGAAAGGEAAATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGV 228
E++ +GA + + TF + L + A ++ PT +Q IPLALQQ
Sbjct 4 EKKSVEGAYI--DDSTTFEAFHLDSRLLQAIKNIGFQYPTLIQSHAIPLALQQ------- 54
Query 229 RTVEQRDVIAVAATGSGKTXAFVLPLLQHLLQHGSSSSSSSSS 271
+RD+IA AATGSGKT A+++P+++ +L++ + + +
Sbjct 55 ----KRDIIAKAATGSGKTLAYLIPVIETILEYKKTIDNGEEN 93
> ath:AT4G34910 DEAD/DEAH box helicase, putative (RH16); K14810
ATP-dependent RNA helicase DDX56/DBP9 [EC:3.6.4.13]
Length=626
Score = 57.8 bits (138), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 32/78 (41%), Positives = 40/78 (51%), Gaps = 12/78 (15%)
Query 182 AAATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAA 241
A +F LGL + A E+PT +QQ IP L E +DV+A A
Sbjct 44 APKSFEELGLDSRLIRALTKKGIEKPTLIQQSAIPYIL------------EGKDVVARAK 91
Query 242 TGSGKTXAFVLPLLQHLL 259
TGSGKT A++LPLLQ L
Sbjct 92 TGSGKTLAYLLPLLQKLF 109
> cel:Y71G12B.8 hypothetical protein; K13181 ATP-dependent RNA
helicase DDX27 [EC:3.6.4.13]
Length=763
Score = 57.8 bits (138), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 30/86 (34%), Positives = 46/86 (53%), Gaps = 12/86 (13%)
Query 174 DGAAAGGEAAATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQ 233
DG + +F + L R++ A + A + PT +QQ IP+AL
Sbjct 138 DGKSLDTSVNVSFEQMNLSRQILKACSGAGYSDPTPIQQACIPVALTG------------ 185
Query 234 RDVIAVAATGSGKTXAFVLPLLQHLL 259
+D+ A AATG+GKT AFVLP+L+ ++
Sbjct 186 KDICACAATGTGKTAAFVLPILERMI 211
> tgo:TGME49_112990 ATP-dependent RNA helicase, putative (EC:5.99.1.3);
K13179 ATP-dependent RNA helicase DDX18/HAS1 [EC:3.6.4.13]
Length=569
Score = 57.4 bits (137), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 42/116 (36%), Positives = 58/116 (50%), Gaps = 21/116 (18%)
Query 154 SSSSAPEAQGVEQQQ----EQQ--QQDGAAAGGEAA---ATFASLGLCREMCAAAAAAKW 204
SS+ PE++G EQQ Q+ A G ++ TF SL +C + A A K
Sbjct 46 SSTEEPESEGPTDDSGDSAEQQGVQKTDKATGKDSFFSDVTFESLDICDPVKKALAEMKM 105
Query 205 ERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAATGSGKTXAFVLPLLQHLLQ 260
ER T +Q + IP R +E RDV+ A TGSGKT AF++P ++ L Q
Sbjct 106 ERLTEIQAKSIP------------RLLEGRDVLGAAKTGSGKTLAFLVPAVELLYQ 149
> ath:AT2G33730 DEAD box RNA helicase, putative; K12858 ATP-dependent
RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=733
Score = 57.4 bits (137), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 29/68 (42%), Positives = 39/68 (57%), Gaps = 12/68 (17%)
Query 191 LCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAATGSGKTXAF 250
L E+ A A +++P+ +Q IPL LQQ RDVI +A TGSGKT AF
Sbjct 320 LTSELLKAVERAGYKKPSPIQMAAIPLGLQQ------------RDVIGIAETGSGKTAAF 367
Query 251 VLPLLQHL 258
VLP+L ++
Sbjct 368 VLPMLAYI 375
> tpv:TP04_0265 small nuclear ribonucleoprotein; K12858 ATP-dependent
RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=744
Score = 57.4 bits (137), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 31/74 (41%), Positives = 42/74 (56%), Gaps = 12/74 (16%)
Query 185 TFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAATGS 244
T+A L E+ A A + +PT +Q Q IP+AL E RD+I +A TGS
Sbjct 330 TWAESPLPWELLEAIKKAGYIKPTPIQMQAIPIAL------------EMRDLIGIAVTGS 377
Query 245 GKTXAFVLPLLQHL 258
GKT AFVLP+L ++
Sbjct 378 GKTAAFVLPMLTYV 391
> mmu:52513 Ddx56, 2600001H07Rik, D11Ertd619e, Noh61; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 56 (EC:3.6.4.13); K14810 ATP-dependent
RNA helicase DDX56/DBP9 [EC:3.6.4.13]
Length=546
Score = 57.4 bits (137), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 31/80 (38%), Positives = 43/80 (53%), Gaps = 12/80 (15%)
Query 181 EAAATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVA 240
+ A F +GL + A W RPT +Q++ IPLAL E +D++A A
Sbjct 4 QEALGFEHMGLDPRLLQAVTDLGWSRPTLIQEKAIPLAL------------EGKDLLARA 51
Query 241 ATGSGKTXAFVLPLLQHLLQ 260
TGSGKT A+ +P+LQ LL
Sbjct 52 RTGSGKTAAYAIPMLQSLLH 71
> dre:445399 ddx56, noh61, zgc:110078; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 56 (EC:3.6.4.13); K14810 ATP-dependent RNA helicase
DDX56/DBP9 [EC:3.6.4.13]
Length=557
Score = 57.0 bits (136), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 30/79 (37%), Positives = 44/79 (55%), Gaps = 12/79 (15%)
Query 181 EAAATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVA 240
E + F +GL + A A W +PT +Q++ IPLAL E +D++A A
Sbjct 3 EDSVRFHEMGLDDRLLKALADLGWSQPTLIQEKAIPLAL------------EGKDLLARA 50
Query 241 ATGSGKTXAFVLPLLQHLL 259
TGSGKT A+ +PL+Q +L
Sbjct 51 RTGSGKTAAYAVPLIQRVL 69
> pfa:PFL2475w DEAD/DEAH box helicase, putative; K13181 ATP-dependent
RNA helicase DDX27 [EC:3.6.4.13]
Length=717
Score = 57.0 bits (136), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 29/80 (36%), Positives = 42/80 (52%), Gaps = 12/80 (15%)
Query 181 EAAATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVA 240
+ ++ L + R K+ PT +Q+ VIPLAL E + ++A +
Sbjct 84 DMNCLWSDLYISRPFLKVLYEQKFSNPTYIQRDVIPLAL------------EGKSILANS 131
Query 241 ATGSGKTXAFVLPLLQHLLQ 260
TGSGKT AFVLP+L+ LLQ
Sbjct 132 ETGSGKTLAFVLPILERLLQ 151
> hsa:54606 DDX56, DDX21, DDX26, NOH61; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 56 (EC:3.6.4.13); K14810 ATP-dependent RNA
helicase DDX56/DBP9 [EC:3.6.4.13]
Length=547
Score = 57.0 bits (136), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 31/78 (39%), Positives = 42/78 (53%), Gaps = 12/78 (15%)
Query 183 AATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAAT 242
A F +GL + A W RPT +Q++ IPLAL E +D++A A T
Sbjct 6 ALGFEHMGLDPRLLQAVTDLGWSRPTLIQEKAIPLAL------------EGKDLLARART 53
Query 243 GSGKTXAFVLPLLQHLLQ 260
GSGKT A+ +P+LQ LL
Sbjct 54 GSGKTAAYAIPMLQLLLH 71
> hsa:79039 DDX54, DP97, MGC2835; DEAD (Asp-Glu-Ala-Asp) box polypeptide
54 (EC:3.6.4.13); K14808 ATP-dependent RNA helicase
DDX54/DBP10 [EC:3.6.4.13]
Length=882
Score = 56.6 bits (135), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 31/111 (27%), Positives = 57/111 (51%), Gaps = 20/111 (18%)
Query 151 ASCSSSSAPEAQGVEQQQEQQQQDGAAAGGEAAATFASLGLCREMCAAAAAAKWERPTAV 210
+ C+S P+ + + + Q ++++ +GG F S+GL + ++ PT +
Sbjct 71 SECTSDVEPDTREMVRAQNKKKK---KSGG-----FQSMGLSYPVFKGIMKKGYKVPTPI 122
Query 211 QQQVIPLALQQLQRLRGVRTVEQRDVIAVAATGSGKTXAFVLPLLQHLLQH 261
Q++ IP+ L + +DV+A+A TGSGKT F+LP+ + L H
Sbjct 123 QRKTIPVIL------------DGKDVVAMARTGSGKTACFLLPMFERLKTH 161
> eco:b3162 deaD, csdA, ECK3150, JW5531, mssB, rhlD; ATP-dependent
RNA helicase; K05592 ATP-dependent RNA helicase DeaD [EC:3.6.4.13]
Length=629
Score = 56.2 bits (134), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 32/78 (41%), Positives = 41/78 (52%), Gaps = 12/78 (15%)
Query 181 EAAATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVA 240
E TFA LGL + A +E+P+ +Q + IP L RDV+ +A
Sbjct 3 EFETTFADLGLKAPILEALNDLGYEKPSPIQAECIPHLLNG------------RDVLGMA 50
Query 241 ATGSGKTXAFVLPLLQHL 258
TGSGKT AF LPLLQ+L
Sbjct 51 QTGSGKTAAFSLPLLQNL 68
> xla:444634 MGC84147 protein
Length=450
Score = 55.8 bits (133), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 26/76 (34%), Positives = 42/76 (55%), Gaps = 12/76 (15%)
Query 184 ATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAATG 243
++FA G ++ +++ +PT +Q Q IP+AL RD+I +A TG
Sbjct 251 SSFAHFGFDEQLMHQIRKSEYTKPTPIQCQGIPVALSG------------RDMIGIAKTG 298
Query 244 SGKTXAFVLPLLQHLL 259
SGKT AF+ P+L H++
Sbjct 299 SGKTAAFIWPILVHIM 314
> cel:C24H12.4 hypothetical protein; K14810 ATP-dependent RNA
helicase DDX56/DBP9 [EC:3.6.4.13]
Length=634
Score = 55.5 bits (132), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 29/84 (34%), Positives = 43/84 (51%), Gaps = 12/84 (14%)
Query 185 TFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAATGS 244
TFA GL + + WE+ VQ+ VI LAL E ++++ A TGS
Sbjct 90 TFADFGLDERILKSIGELGWEKANQVQESVISLAL------------ENKNIMGRARTGS 137
Query 245 GKTXAFVLPLLQHLLQHGSSSSSS 268
GKT AF++PL+Q L+ ++ S
Sbjct 138 GKTGAFLIPLVQKLIAESKTNDGS 161
> bbo:BBOV_I003430 19.m02227; DEAD/DEAH box helicase and helicase
conserved C-terminal domain containing protein; K14808 ATP-dependent
RNA helicase DDX54/DBP10 [EC:3.6.4.13]
Length=783
Score = 55.5 bits (132), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 35/101 (34%), Positives = 53/101 (52%), Gaps = 17/101 (16%)
Query 164 VEQQQE--QQQQDGAAAGGEAAATFASLGLCREMCAAAA-AAKWERPTAVQQQVIPLALQ 220
VE E +Q++ GG A F LGL R +C A ++++P+ +Q++ IP LQ
Sbjct 2 VEDSNENTKQKKKSNHEGGTGA--FGLLGLDRTLCYALEHKLRYKQPSTIQRRTIPAVLQ 59
Query 221 QLQRLRGVRTVEQRDVIAVAATGSGKTXAFVLPLLQHLLQH 261
RDV+ +A TGSGKT A++ P++Q L H
Sbjct 60 G------------RDVVCIARTGSGKTAAYLAPVVQLLEGH 88
> dre:30263 vasa, MGC158535, fi24g05, vas, vlg, wu:fi24g05, zgc:109812,
zgc:158535; vasa homolog (EC:3.6.4.13); K13982 probable
ATP-dependent RNA helicase DDX4 [EC:3.6.4.13]
Length=716
Score = 55.5 bits (132), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 31/89 (34%), Positives = 46/89 (51%), Gaps = 12/89 (13%)
Query 181 EAAATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVA 240
+A TF GLC + + + + +PT VQ+ IP+ RD++A A
Sbjct 274 KAIMTFEEAGLCDSLSKNVSKSGYVKPTPVQKHGIPI------------ISAGRDLMACA 321
Query 241 ATGSGKTXAFVLPLLQHLLQHGSSSSSSS 269
TGSGKT AF+LP+LQ + G ++S S
Sbjct 322 QTGSGKTAAFLLPILQRFMTDGVAASKFS 350
> bbo:BBOV_III008730 17.m07763; DEAD/DEAH box helicase domain
containing protein; K13181 ATP-dependent RNA helicase DDX27
[EC:3.6.4.13]
Length=649
Score = 55.5 bits (132), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 27/75 (36%), Positives = 43/75 (57%), Gaps = 12/75 (16%)
Query 185 TFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAATGS 244
++ LGL R + A ++ P+ +Q +VIP+AL E +D++A A TGS
Sbjct 126 NWSDLGLSRSLIKAVFDMGYKAPSIIQSKVIPVAL------------EGKDLLATAETGS 173
Query 245 GKTXAFVLPLLQHLL 259
GK+ AF++P LQ L+
Sbjct 174 GKSAAFLIPTLQRLI 188
> sce:YHR065C RRP3; Protein involved in rRNA processing; required
for maturation of the 35S primary transcript of pre-rRNA
and for cleavage leading to mature 18S rRNA; homologous to
eIF-4a, which is a DEAD box RNA-dependent ATPase with helicase
activity (EC:3.6.1.-); K14777 ATP-dependent RNA helicase
DDX47/RRP3 [EC:3.6.4.13]
Length=501
Score = 55.1 bits (131), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 32/95 (33%), Positives = 45/95 (47%), Gaps = 18/95 (18%)
Query 164 VEQQQEQQQQDGAAAGGEAAATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQ 223
V Q E +D E+ +F+ L L E+ A + +PT +Q + IP AL
Sbjct 67 VSTQNENTNED------ESFESFSELNLVPELIQACKNLNYSKPTPIQSKAIPPAL---- 116
Query 224 RLRGVRTVEQRDVIAVAATGSGKTXAFVLPLLQHL 258
E D+I +A TGSGKT AF +P+L L
Sbjct 117 --------EGHDIIGLAQTGSGKTAAFAIPILNRL 143
> cel:F01F1.7 ddx-23; DEAD boX helicase homolog family member
(ddx-23); K12858 ATP-dependent RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=730
Score = 55.1 bits (131), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 29/66 (43%), Positives = 35/66 (53%), Gaps = 12/66 (18%)
Query 190 GLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAATGSGKTXA 249
G E+ A + PT +Q+Q IP+ LQ RDVI VA TGSGKT A
Sbjct 307 GFPDEVYQAVKEIGYLEPTPIQRQAIPIGLQN------------RDVIGVAETGSGKTAA 354
Query 250 FVLPLL 255
F+LPLL
Sbjct 355 FLLPLL 360
> mmu:13206 Ddx4, AV206478, Mvh, VASA; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 4 (EC:3.6.4.13); K13982 probable ATP-dependent
RNA helicase DDX4 [EC:3.6.4.13]
Length=728
Score = 55.1 bits (131), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 31/85 (36%), Positives = 46/85 (54%), Gaps = 12/85 (14%)
Query 182 AAATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAA 241
A TF LC+ + A A + + T VQ+ IP+ L RD++A A
Sbjct 285 AILTFEEANLCQTLNNNIAKAGYTKLTPVQKYSIPIVLAG------------RDLMACAQ 332
Query 242 TGSGKTXAFVLPLLQHLLQHGSSSS 266
TGSGKT AF+LP+L H+++ G ++S
Sbjct 333 TGSGKTAAFLLPILAHMMRDGITAS 357
> hsa:54514 DDX4, MGC111074, VASA; DEAD (Asp-Glu-Ala-Asp) box
polypeptide 4 (EC:3.6.4.13); K13982 probable ATP-dependent RNA
helicase DDX4 [EC:3.6.4.13]
Length=690
Score = 54.7 bits (130), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 31/85 (36%), Positives = 45/85 (52%), Gaps = 12/85 (14%)
Query 182 AAATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAA 241
A TF LC+ + A A + + T VQ+ IP+ L RD++A A
Sbjct 252 AILTFEEANLCQTLNNNIAKAGYTKLTPVQKYSIPIILAG------------RDLMACAQ 299
Query 242 TGSGKTXAFVLPLLQHLLQHGSSSS 266
TGSGKT AF+LP+L H++ G ++S
Sbjct 300 TGSGKTAAFLLPILAHMMHDGITAS 324
> dre:503932 ddx42, im:7148194, zgc:111815; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 42 (EC:3.6.1.-); K12835 ATP-dependent RNA
helicase DDX42 [EC:3.6.4.13]
Length=908
Score = 54.7 bits (130), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 41/78 (52%), Gaps = 12/78 (15%)
Query 183 AATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAAT 242
A +FA G ++ +++ +PT +Q Q +P+AL RD I +A T
Sbjct 254 ATSFAHFGFDEQLMHQIRKSEYTQPTPIQCQGVPIALSG------------RDAIGIAKT 301
Query 243 GSGKTXAFVLPLLQHLLQ 260
GSGKT AF+ P+L H++
Sbjct 302 GSGKTAAFIWPILVHIMD 319
> cpv:cgd6_4830 Drs1p, eIF4a-1-family RNA SFII helicase ; K13181
ATP-dependent RNA helicase DDX27 [EC:3.6.4.13]
Length=573
Score = 54.3 bits (129), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 35/98 (35%), Positives = 50/98 (51%), Gaps = 12/98 (12%)
Query 174 DGAAAGGEAAATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQ 233
+G E ++SL L R + A + + T +Q++VIPLAL
Sbjct 20 EGEIKRREKIKMWSSLELSRPLLKALSDLNFVEATLIQKEVIPLALSG------------ 67
Query 234 RDVIAVAATGSGKTXAFVLPLLQHLLQHGSSSSSSSSS 271
RD++A A TGSGKT AF+LP L+ LL+ +S SS
Sbjct 68 RDIMAEAETGSGKTAAFLLPALERLLRSPYVRNSRVSS 105
> sce:YLL008W DRS1; Drs1p (EC:3.6.1.-); K13181 ATP-dependent RNA
helicase DDX27 [EC:3.6.4.13]
Length=752
Score = 53.9 bits (128), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 32/102 (31%), Positives = 51/102 (50%), Gaps = 24/102 (23%)
Query 158 APEAQGVEQQQEQQQQDGAAAGGEAAATFASLGLCREMCAAAAAAKWERPTAVQQQVIPL 217
APE +G E +++ + F SL L R + A+ + +P+ +Q IP+
Sbjct 217 APETEGDEAKKQMYE------------NFNSLSLSRPVLKGLASLGYVKPSPIQSATIPI 264
Query 218 ALQQLQRLRGVRTVEQRDVIAVAATGSGKTXAFVLPLLQHLL 259
AL +D+IA A TGSGKT AF++P+++ LL
Sbjct 265 ALLG------------KDIIAGAVTGSGKTAAFMIPIIERLL 294
> xla:399080 ddx6, p54h; DEAD (Asp-Glu-Ala-Asp) box polypeptide
6 (EC:3.6.4.13); K12614 ATP-dependent RNA helicase DDX6/DHH1
[EC:3.6.4.13]
Length=481
Score = 53.5 bits (127), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 39/73 (53%), Gaps = 12/73 (16%)
Query 186 FASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAATGSG 245
F L RE+ WE+P+ +Q++ IP+AL RD++A A G+G
Sbjct 97 FEDYCLKRELLMGIFEMGWEKPSPIQEESIPIALSG------------RDILARAKNGTG 144
Query 246 KTXAFVLPLLQHL 258
KT A+++PLL+ L
Sbjct 145 KTGAYLIPLLERL 157
> mmu:71990 Ddx54, 2410015A15Rik, AI414901, DP97; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 54 (EC:3.6.4.13); K14808 ATP-dependent
RNA helicase DDX54/DBP10 [EC:3.6.4.13]
Length=874
Score = 53.5 bits (127), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 30/108 (27%), Positives = 55/108 (50%), Gaps = 20/108 (18%)
Query 151 ASCSSSSAPEAQGVEQQQEQQQQDGAAAGGEAAATFASLGLCREMCAAAAAAKWERPTAV 210
+ C S P+ + + + Q ++++ +GG F S+GL + ++ PT +
Sbjct 70 SECVSDVEPDTREMVRAQNKKKK---KSGG-----FQSMGLSYPVFKGIMKKGYKVPTPI 121
Query 211 QQQVIPLALQQLQRLRGVRTVEQRDVIAVAATGSGKTXAFVLPLLQHL 258
Q++ IP+ L + +DV+A+A TGSGKT F+LP+ + L
Sbjct 122 QRKTIPVIL------------DGKDVVAMARTGSGKTACFLLPMFERL 157
> dre:386632 ddx49, wu:fb82g04, wu:fd12e05; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 49 (EC:3.6.4.13); K14778 ATP-dependent
RNA helicase DDX49/DBP8 [EC:3.6.4.13]
Length=468
Score = 53.5 bits (127), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 33/76 (43%), Positives = 39/76 (51%), Gaps = 12/76 (15%)
Query 183 AATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAAT 242
ATF SLGL + RPTAVQ++ IP L + RD + A T
Sbjct 1 MATFESLGLSEWLIQQCKQMGISRPTAVQEKCIPAIL------------DGRDCMGCAKT 48
Query 243 GSGKTXAFVLPLLQHL 258
GSGKT AFVLP+LQ L
Sbjct 49 GSGKTAAFVLPVLQKL 64
> bbo:BBOV_II003280 18.m06276; DEAD box RNA helicase; K12858 ATP-dependent
RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=714
Score = 53.1 bits (126), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 30/74 (40%), Positives = 42/74 (56%), Gaps = 12/74 (16%)
Query 185 TFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAATGS 244
T+A L E+ A A ++ PT +Q Q IP+ L G+R D+I +A TGS
Sbjct 298 TWAESNLPSELLRAIKDAGFKSPTPIQMQAIPIGL-------GMR-----DLIGLAETGS 345
Query 245 GKTXAFVLPLLQHL 258
GKT AFVLP+L ++
Sbjct 346 GKTVAFVLPMLTYV 359
> pfa:PFL1310c DEAD-box helicase; K12823 ATP-dependent RNA helicase
DDX5/DBP2 [EC:3.6.4.13]
Length=742
Score = 52.4 bits (124), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 26/59 (44%), Positives = 34/59 (57%), Gaps = 12/59 (20%)
Query 203 KWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAATGSGKTXAFVLPLLQHLLQH 261
K+ PTA+Q+ P+AL +D+I VA TGSGKT AFVLP H+L+H
Sbjct 306 KFSEPTAIQKITWPIALSG------------KDLIGVAETGSGKTLAFVLPCFMHILKH 352
> ath:AT4G16630 DEAD/DEAH box helicase, putative (RH28); K13181
ATP-dependent RNA helicase DDX27 [EC:3.6.4.13]
Length=789
Score = 52.4 bits (124), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 32/77 (41%), Positives = 39/77 (50%), Gaps = 12/77 (15%)
Query 183 AATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAAT 242
A TF L L R + A +++PT +Q IPLAL RD+ A A T
Sbjct 166 ADTFMELNLSRPLLRACETLGYKKPTPIQAACIPLALTG------------RDLCASAIT 213
Query 243 GSGKTXAFVLPLLQHLL 259
GSGKT AF LP L+ LL
Sbjct 214 GSGKTAAFALPTLERLL 230
> sce:YDL031W DBP10; Dbp10p (EC:3.6.1.-); K14808 ATP-dependent
RNA helicase DDX54/DBP10 [EC:3.6.4.13]
Length=995
Score = 52.4 bits (124), Expect = 2e-06, Method: Composition-based stats.
Identities = 27/78 (34%), Positives = 42/78 (53%), Gaps = 12/78 (15%)
Query 184 ATFASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAATG 243
+F S GL + + + +PT +Q++ IPL LQ RD++ +A TG
Sbjct 137 GSFPSFGLSKIVLNNIKRKGFRQPTPIQRKTIPLILQS------------RDIVGMARTG 184
Query 244 SGKTXAFVLPLLQHLLQH 261
SGKT AF+LP+++ L H
Sbjct 185 SGKTAAFILPMVEKLKSH 202
> mmu:13209 Ddx6, 1110001P04Rik, E230023J21Rik, HLR2, mRCK/P54,
p54, rck; DEAD (Asp-Glu-Ala-Asp) box polypeptide 6 (EC:3.6.4.13);
K12614 ATP-dependent RNA helicase DDX6/DHH1 [EC:3.6.4.13]
Length=483
Score = 52.4 bits (124), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 24/73 (32%), Positives = 39/73 (53%), Gaps = 12/73 (16%)
Query 186 FASLGLCREMCAAAAAAKWERPTAVQQQVIPLALQQLQRLRGVRTVEQRDVIAVAATGSG 245
F L RE+ WE+P+ +Q++ IP+AL RD++A A G+G
Sbjct 98 FEDYCLKRELLMGIFEMGWEKPSPIQEESIPIAL------------SGRDILARAKNGTG 145
Query 246 KTXAFVLPLLQHL 258
K+ A+++PLL+ L
Sbjct 146 KSGAYLIPLLERL 158
Lambda K H
0.310 0.114 0.302
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 10209973120
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40