bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0113_orf3
Length=329
Score E
Sequences producing significant alignments: (Bits) Value
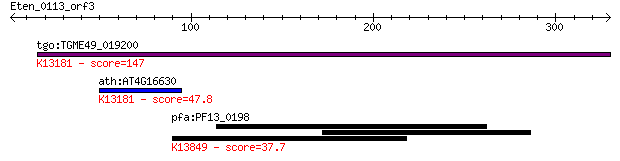
tgo:TGME49_019200 DEAD/DEAH box RNA helicase, putative ; K1318... 147 7e-35
ath:AT4G16630 DEAD/DEAH box helicase, putative (RH28); K13181 ... 47.8 6e-05
pfa:PF13_0198 PfRh2a; reticulocyte binding protein 2 homolog A... 37.7 0.058
> tgo:TGME49_019200 DEAD/DEAH box RNA helicase, putative ; K13181
ATP-dependent RNA helicase DDX27 [EC:3.6.4.13]
Length=962
Score = 147 bits (370), Expect = 7e-35, Method: Compositional matrix adjust.
Identities = 116/327 (35%), Positives = 190/327 (58%), Gaps = 36/327 (11%)
Query 16 VLRRRIDSANLEAIHKELRGLEGALQKKRQEETLEREMRLAEIFIAKANNIQQHADEIYS 75
V +RRID+ LE + K+++ L+G + ++ + E LERE+RLAE+ + KA N+Q HADEIYS
Sbjct 659 VFQRRIDADKLEMLEKKVKSLQGDIVRELKREKLEREVRLAELHLQKAENLQTHADEIYS 718
Query 76 RPHREWFVSAGDKRKLQEASRI----DAEARALISTSSSTDSKEATKDTKQKQKHRQQQQ 131
RP R+WF++A +K++L++ S+ DAE R + + ++ S E ++ + + ++
Sbjct 719 RPMRQWFMTAKEKQRLKDESKALVGKDAEERDALERARASSSDE--RNGRAPLSEDEAEE 776
Query 132 QDTTESDSSDAADSSSGGEEEEESEE------ELPEWATAESVGSEEEEEDLEGAEKTSS 185
TE+D++D DS +EE+ ++E ELPEW TA S S+ E+++E
Sbjct 777 DSETEADNADQMDSD---DEEDVADESNSDDGELPEWITACSDDSQSVEDEIE------- 826
Query 186 SEPKALVKVKEIKVTKEQKARLQQQQQQLQKKEKKHQTMTPRQKERMQEFRMAKAAARSV 245
EP K K + V + + + + Q K++KK + +TP+Q+ERM+E + KAAARS
Sbjct 827 -EPPPKKKAKAVTVKEFKATKPKAQPIGGNKEKKKQERLTPKQRERMKELQFMKAAARSA 885
Query 246 KKNARPQRLRIDTEVDEDYLGGSRRPRSRKPQKKKR---RREEIERSSGNNSKVSGVDKS 302
K++ P+RLR+ D SRRP+++K +K K R EE ++ G ++
Sbjct 886 KRSQMPKRLRVTHTSPNDVHNNSRRPKNQKKRKLKASGWRAEEPKKPQGGKNE------- 938
Query 303 VGPAQKKRRGTPVGSGSFKSKKRYKRR 329
+K+R +G +FKSK RYKRR
Sbjct 939 ---NKKQRNVKTMGKKAFKSKGRYKRR 962
> ath:AT4G16630 DEAD/DEAH box helicase, putative (RH28); K13181
ATP-dependent RNA helicase DDX27 [EC:3.6.4.13]
Length=789
Score = 47.8 bits (112), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 21/45 (46%), Positives = 32/45 (71%), Gaps = 0/45 (0%)
Query 50 EREMRLAEIFIAKANNIQQHADEIYSRPHREWFVSAGDKRKLQEA 94
ER +R AE+ AKA N+ +H DEIY+RP R WF++ +K+ + +A
Sbjct 569 ERALRKAEMEFAKAENMLEHRDEIYARPKRTWFMTEKEKKLVAQA 613
> pfa:PF13_0198 PfRh2a; reticulocyte binding protein 2 homolog
A; K13849 reticulocyte-binding protein
Length=3130
Score = 37.7 bits (86), Expect = 0.058, Method: Composition-based stats.
Identities = 32/150 (21%), Positives = 76/150 (50%), Gaps = 9/150 (6%)
Query 114 KEATKDTKQKQKHRQQQQQDTTESDSSDAADSSSGGEEEEESEEELPEWATAESVGSEEE 173
K ++ QK++ ++Q+Q+ E + + ++EEE + + E E +E
Sbjct 2740 KRQEQERLQKEEELKRQEQERLEREKQEQL------QKEEELKRQEQERLQKEEALKRQE 2793
Query 174 EEDLEGAEKTSSSEPKALVKVKEIKVTKEQKARLQQQQQQLQKKE--KKHQTMTPRQKER 231
+E L+ E+ E + L + K+ ++ KE++ + +Q+Q++LQK+E K+ + +++E
Sbjct 2794 QERLQKEEELKRQEQERLEREKQEQLQKEEELK-RQEQERLQKEEALKRQEQERLQKEEE 2852
Query 232 MQEFRMAKAAARSVKKNARPQRLRIDTEVD 261
++ + + ++ R Q ++ E D
Sbjct 2853 LKRQEQERLERKKIELAEREQHIKSKLESD 2882
Score = 35.0 bits (79), Expect = 0.38, Method: Composition-based stats.
Identities = 27/118 (22%), Positives = 60/118 (50%), Gaps = 4/118 (3%)
Query 172 EEEEDLEGAEKTSSSEPKALVKVKEIKVTKEQKARLQQQQQQLQKKEKKHQTMTPRQKE- 230
+E+E L+ E+ E + L + K+ ++ KE++ + Q+Q++ +++ K Q QKE
Sbjct 2742 QEQERLQKEEELKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQKEE 2801
Query 231 ---RMQEFRMAKAAARSVKKNARPQRLRIDTEVDEDYLGGSRRPRSRKPQKKKRRREE 285
R ++ R+ + ++K +R + E+ L + R +K ++ KR+ +E
Sbjct 2802 ELKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQKEEELKRQEQE 2859
Score = 32.3 bits (72), Expect = 2.4, Method: Composition-based stats.
Identities = 26/128 (20%), Positives = 63/128 (49%), Gaps = 2/128 (1%)
Query 90 KLQEASRIDAEARALISTSSSTDSKEATKDTKQKQKHRQQQQQDTTESDSSDAADSSSGG 149
K QE R+ E + + ++ QK++ ++Q+Q+ + + +
Sbjct 2740 KRQEQERLQKEEE--LKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERL 2797
Query 150 EEEEESEEELPEWATAESVGSEEEEEDLEGAEKTSSSEPKALVKVKEIKVTKEQKARLQQ 209
++EEE + + E E ++EE+L+ E+ + +AL + ++ ++ KE++ + Q+
Sbjct 2798 QKEEELKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQKEEELKRQE 2857
Query 210 QQQQLQKK 217
Q++ +KK
Sbjct 2858 QERLERKK 2865
Lambda K H
0.304 0.119 0.308
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 13712401440
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40