bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0114_orf1
Length=172
Score E
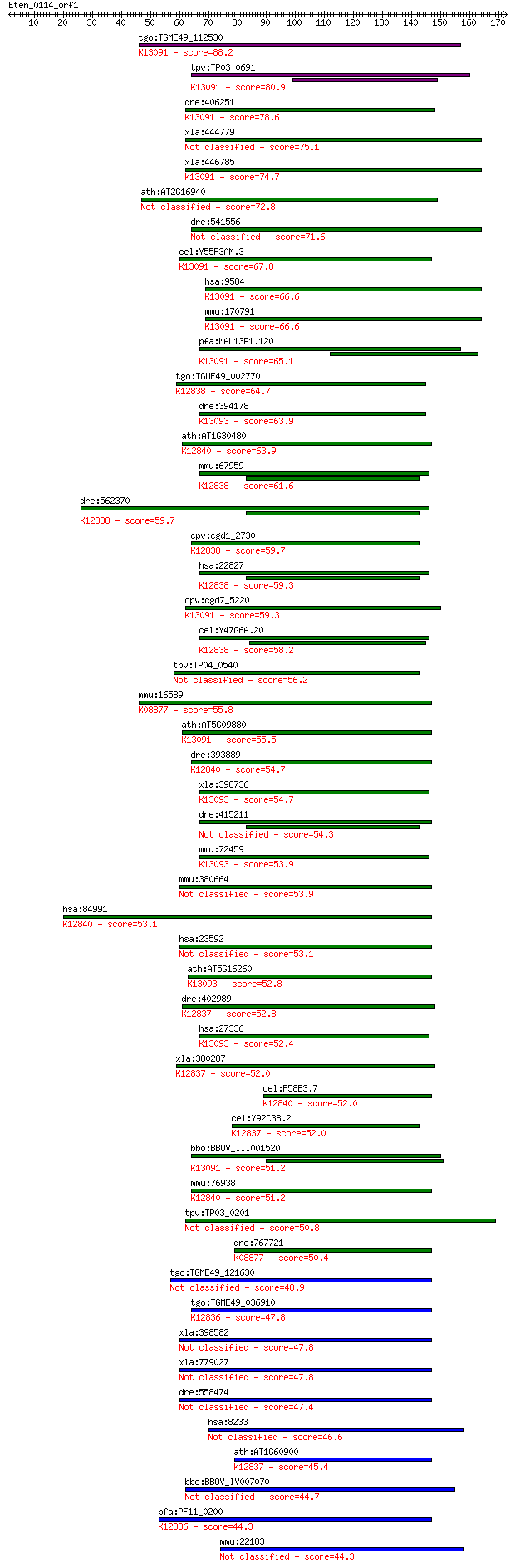
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_112530 splicing factor protein, putative ; K13091 R... 88.2 1e-17
tpv:TP03_0691 splicing factor; K13091 RNA-binding protein 39 80.9
dre:406251 rbm39a, rnpc2, zgc:55780; RNA binding motif protein... 78.6 1e-14
xla:444779 MGC81970 protein 75.1 1e-13
xla:446785 rbm39, MGC80448, rnpc2; RNA binding motif protein 3... 74.7 1e-13
ath:AT2G16940 RNA recognition motif (RRM)-containing protein 72.8 5e-13
dre:541556 rbm39b, rnpc2l, wu:fa97g07, wu:fb09c08, zgc:112139,... 71.6 1e-12
cel:Y55F3AM.3 hypothetical protein; K13091 RNA-binding protein 39 67.8
hsa:9584 RBM39, CAPER, CAPERalpha, CC1.3, DKFZp781C0423, FLJ44... 66.6 4e-11
mmu:170791 Rbm39, 1500012C14Rik, 2310040E03Rik, B330012G18Rik,... 66.6 4e-11
pfa:MAL13P1.120 splicing factor, putative; K13091 RNA-binding ... 65.1 1e-10
tgo:TGME49_002770 RNA-binding protein, putative ; K12838 poly(... 64.7 2e-10
dre:394178 htatsf1, MGC66252, zgc:66252; HIV TAT specific fact... 63.9 3e-10
ath:AT1G30480 DRT111; DRT111; nucleic acid binding / nucleotid... 63.9 3e-10
mmu:67959 Puf60, 2410104I19Rik, 2810454F19Rik, SIAHBP1; poly-U... 61.6 1e-09
dre:562370 puf60a, fc21a05, wu:fc21a05, wu:fi43a08; poly-U bin... 59.7 4e-09
cpv:cgd1_2730 Ro ribonucleoprotein-binding protein 1, RNA bind... 59.7 4e-09
hsa:22827 PUF60, FIR, FLJ31379, RoBPI, SIAHBP1; poly-U binding... 59.3 6e-09
cpv:cgd7_5220 splicing factor with 3 RRM domains ; K13091 RNA-... 59.3 6e-09
cel:Y47G6A.20 rnp-6; RNP (RRM RNA binding domain) containing f... 58.2 1e-08
tpv:TP04_0540 hypothetical protein 56.2 5e-08
mmu:16589 Uhmk1, 4732477C12Rik, 4930500M09Rik, AA673513, AI449... 55.8 6e-08
ath:AT5G09880 RNA recognition motif (RRM)-containing protein; ... 55.5 8e-08
dre:393889 rbm17, MGC55840, zgc:55840; RNA binding motif prote... 54.7 2e-07
xla:398736 htatsf1, MGC98898; HIV-1 Tat specific factor 1; K13... 54.7 2e-07
dre:415211 puf60b, SIAHBP1, fe37c05, repressor, si:ch211-12p8.... 54.3 2e-07
mmu:72459 Htatsf1, 1600023H17Rik, 2600017A12Rik, 2700077B20Rik... 53.9 3e-07
mmu:380664 Lemd3, AI316861, Man1; LEM domain containing 3 53.9
hsa:84991 RBM17, DKFZp686F13131, MGC14439, SPF45; RNA binding ... 53.1 4e-07
hsa:23592 LEMD3, MAN1; LEM domain containing 3 53.1
ath:AT5G16260 RNA recognition motif (RRM)-containing protein; ... 52.8 6e-07
dre:402989 u2af2b, MGC77804, wu:fb73g02, zgc:77804; U2 small n... 52.8 6e-07
hsa:27336 HTATSF1, TAT-SF1, dJ196E23.2; HIV-1 Tat specific fac... 52.4 8e-07
xla:380287 u2af2, MGC53441, u2af65; U2 small nuclear RNA auxil... 52.0 9e-07
cel:F58B3.7 hypothetical protein; K12840 splicing factor 45 52.0
cel:Y92C3B.2 uaf-1; U2AF splicing factor family member (uaf-1)... 52.0 1e-06
bbo:BBOV_III001520 17.m07155; splicing factor, CC1-like family... 51.2 2e-06
mmu:76938 Rbm17, 2700027J02Rik; RNA binding motif protein 17; ... 51.2 2e-06
tpv:TP03_0201 hypothetical protein 50.8 2e-06
dre:767721 uhmk1, MGC153241, zgc:153241; U2AF homology motif (... 50.4 3e-06
tgo:TGME49_121630 hypothetical protein 48.9 9e-06
tgo:TGME49_036910 U2 snRNP auxiliary factor small subunit, put... 47.8 2e-05
xla:398582 lemd3, man1, sane, xman1; LEM domain containing 3 47.8
xla:779027 sane, LEMD3; Smad1 antagonistic effector 47.8 2e-05
dre:558474 lemd3, im:7158446, si:dkey-6a5.6, wu:fb34b12; LEM d... 47.4 3e-05
hsa:8233 ZRSR2, MGC142014, MGC142040, U2AF1-RS2, U2AF1L2, U2AF... 46.6 5e-05
ath:AT1G60900 U2 snRNP auxiliary factor large subunit, putativ... 45.4 8e-05
bbo:BBOV_IV007070 23.m05938; hypothetical protein 44.7 2e-04
pfa:PF11_0200 U2 snRNP auxiliary factor, small subunit, putati... 44.3 2e-04
mmu:22183 Zrsr1, D11Ncvs75, Irlgs2, SP2, U2af1-rs1, U2afbp-rs;... 44.3 2e-04
> tgo:TGME49_112530 splicing factor protein, putative ; K13091
RNA-binding protein 39
Length=633
Score = 88.2 bits (217), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 47/114 (41%), Positives = 72/114 (63%), Gaps = 5/114 (4%)
Query 46 FLRSSSGPRVGGLAPGVMTSNLVLRGMF---NPSAADDEYYFEDIRDDVREECGKHGSVV 102
FL +S+G A G + N+VL MF + + +D ++F D+ DDVR+EC K GSV
Sbjct 521 FLPASNGALFADNATG--SCNVVLHNMFAAKDVNLKEDPHFFLDLGDDVRDECKKFGSVE 578
Query 103 QVSLSEKDEEGRVFVKFTAPSVALAAQNALNGRFFGGRQIHAEFATDAMINAVC 156
+V + E++ +G V+++F P A AA ALNGR+F G+ I AEF +DA+ ++ C
Sbjct 579 KVWIDERNVDGNVWIRFAHPDQARAAFGALNGRYFAGKPISAEFISDAVWSSTC 632
> tpv:TP03_0691 splicing factor; K13091 RNA-binding protein 39
Length=644
Score = 80.9 bits (198), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 37/97 (38%), Positives = 61/97 (62%), Gaps = 1/97 (1%)
Query 64 TSNLVLRGMFNPS-AADDEYYFEDIRDDVREECGKHGSVVQVSLSEKDEEGRVFVKFTAP 122
+SNLVL M+ + D+ +F++I +DV+EECGK+G+V+QV +++++ +G+V+VKF
Sbjct 548 SSNLVLSNMYTSADYEDNREFFDEIEEDVKEECGKYGTVIQVFVNKRNPDGKVYVKFKNN 607
Query 123 SVALAAQNALNGRFFGGRQIHAEFATDAMINAVCGNS 159
A AA +L GR+F G I + +D V S
Sbjct 608 DDAQAANKSLQGRYFAGNTIQVSYISDDQYQDVVNKS 644
Score = 32.0 bits (71), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 15/53 (28%), Positives = 29/53 (54%), Gaps = 3/53 (5%)
Query 99 GSVVQVSLSEKDE---EGRVFVKFTAPSVALAAQNALNGRFFGGRQIHAEFAT 148
G+++ V+L+ D+ +G +++F + A A N +NG G+QI +A
Sbjct 447 GNIIDVALARTDDGNSKGYAYIRFKRWNEAKEALNVMNGFDINGQQIKVAYAN 499
> dre:406251 rbm39a, rnpc2, zgc:55780; RNA binding motif protein
39a; K13091 RNA-binding protein 39
Length=523
Score = 78.6 bits (192), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 36/86 (41%), Positives = 54/86 (62%), Gaps = 0/86 (0%)
Query 62 VMTSNLVLRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSLSEKDEEGRVFVKFTA 121
+ T L MFNP++ +D + +I+DDV EEC KHG V+ + + +K EG V+VK
Sbjct 414 IATHCFQLSNMFNPNSENDHGWEIEIQDDVIEECNKHGGVIHIYVDKKSAEGNVYVKCPT 473
Query 122 PSVALAAQNALNGRFFGGRQIHAEFA 147
A+AA +AL+GR+FGG+ I A +
Sbjct 474 IPAAMAAVSALHGRWFGGKMITAAYV 499
> xla:444779 MGC81970 protein
Length=512
Score = 75.1 bits (183), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 35/102 (34%), Positives = 59/102 (57%), Gaps = 0/102 (0%)
Query 62 VMTSNLVLRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSLSEKDEEGRVFVKFTA 121
+ T L MFNP D+ + +I++DV EEC KHG + + + + +G V+VK +
Sbjct 403 IATQCFQLSNMFNPQTEDELGWDSEIKEDVMEECNKHGGAIHIYVDKNSPQGNVYVKCST 462
Query 122 PSVALAAQNALNGRFFGGRQIHAEFATDAMINAVCGNSSTNT 163
+ A+AA NAL+GR+F G+ I A + +++ +S T+T
Sbjct 463 ITSAIAAVNALHGRWFAGKMITAAYVPVPTYHSLFPDSMTST 504
> xla:446785 rbm39, MGC80448, rnpc2; RNA binding motif protein
39; K13091 RNA-binding protein 39
Length=540
Score = 74.7 bits (182), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 37/102 (36%), Positives = 59/102 (57%), Gaps = 0/102 (0%)
Query 62 VMTSNLVLRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSLSEKDEEGRVFVKFTA 121
+ T L MFNP D+ + +I++DV EEC KHG VV + + + +G V+VK
Sbjct 431 IATQCFQLSNMFNPQTEDELGWDSEIKEDVIEECNKHGGVVHLYVDKNSAQGNVYVKCPT 490
Query 122 PSVALAAQNALNGRFFGGRQIHAEFATDAMINAVCGNSSTNT 163
+ A+AA NAL+GR+F G+ I A + +++ +S T+T
Sbjct 491 IASAIAAVNALHGRWFAGKMITAAYVPLPTYHSLFPDSMTST 532
> ath:AT2G16940 RNA recognition motif (RRM)-containing protein
Length=561
Score = 72.8 bits (177), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 42/117 (35%), Positives = 65/117 (55%), Gaps = 16/117 (13%)
Query 47 LRSSSGPRVGGLA-----PGVMTSN----------LVLRGMFNPSAADDEYYFEDIRDDV 91
L S P V GLA PGV+ + L+L+ MF+PS ++ + EDI++DV
Sbjct 432 LVQGSFPAVAGLAGSGIIPGVIPAGFDPIGVPSECLLLKNMFDPSTETEDDFDEDIKEDV 491
Query 92 REECGKHGSVVQVSLSEKDEEGRVFVKFTAPSVALAAQNALNGRFFGGRQIHAEFAT 148
+EEC K G + + +K+ G V+++F A+ AQ AL+GR+F G+ I A + T
Sbjct 492 KEECSKFGKLNHI-FVDKNSVGFVYLRFENAQAAIGAQRALHGRWFAGKMITATYMT 547
> dre:541556 rbm39b, rnpc2l, wu:fa97g07, wu:fb09c08, zgc:112139,
zgc:113117; RNA binding motif protein 39b
Length=539
Score = 71.6 bits (174), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 35/100 (35%), Positives = 57/100 (57%), Gaps = 0/100 (0%)
Query 64 TSNLVLRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSLSEKDEEGRVFVKFTAPS 123
T L L MFNP ++ + +IRDDV EEC KHG V+ + + + +G V+VK
Sbjct 432 THCLQLSNMFNPQMENEPGWDIEIRDDVIEECRKHGGVIHIYVDKNSAQGNVYVKCPTIP 491
Query 124 VALAAQNALNGRFFGGRQIHAEFATDAMINAVCGNSSTNT 163
VA+A ++L+GR+F G+ I A + + + +++T T
Sbjct 492 VAMAVVSSLHGRWFAGKMITAAYVPLPTYHNLFPDAATAT 531
> cel:Y55F3AM.3 hypothetical protein; K13091 RNA-binding protein
39
Length=580
Score = 67.8 bits (164), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 34/89 (38%), Positives = 53/89 (59%), Gaps = 4/89 (4%)
Query 60 PGVMTSNLVLRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSLSEKDEEGRVFVKF 119
P + T +L MF+PS + + DIR+DV E+C HG + V + + E+G V+VK
Sbjct 417 PSIATQCFLLSNMFDPSKETEPAWDHDIREDVIEQCLAHGGALHVFVDKGSEQGNVYVK- 475
Query 120 TAPSVALAAQ--NALNGRFFGGRQIHAEF 146
PS+ +A Q +AL+GR+F G+ I A +
Sbjct 476 -CPSIVIAHQAVSALHGRWFSGKVITANY 503
> hsa:9584 RBM39, CAPER, CAPERalpha, CC1.3, DKFZp781C0423, FLJ44170,
HCC1, RNPC2, fSAP59; RNA binding motif protein 39; K13091
RNA-binding protein 39
Length=524
Score = 66.6 bits (161), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 35/95 (36%), Positives = 56/95 (58%), Gaps = 0/95 (0%)
Query 69 LRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSLSEKDEEGRVFVKFTAPSVALAA 128
L MFNP ++ + +I+DDV EEC KHG V+ + + + +G V+VK + + A+AA
Sbjct 422 LSNMFNPQTEEEVGWDTEIKDDVIEECNKHGGVIHIYVDKNSAQGNVYVKCPSIAAAIAA 481
Query 129 QNALNGRFFGGRQIHAEFATDAMINAVCGNSSTNT 163
NAL+GR+F G+ I A + + + +S T T
Sbjct 482 VNALHGRWFAGKMITAAYVPLPTYHNLFPDSMTAT 516
> mmu:170791 Rbm39, 1500012C14Rik, 2310040E03Rik, B330012G18Rik,
C79248, R75070, Rnpc2, caper; RNA binding motif protein 39;
K13091 RNA-binding protein 39
Length=530
Score = 66.6 bits (161), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 35/95 (36%), Positives = 56/95 (58%), Gaps = 0/95 (0%)
Query 69 LRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSLSEKDEEGRVFVKFTAPSVALAA 128
L MFNP ++ + +I+DDV EEC KHG V+ + + + +G V+VK + + A+AA
Sbjct 428 LSNMFNPQTEEEVGWDTEIKDDVIEECNKHGGVIHIYVDKNSAQGNVYVKCPSIAAAIAA 487
Query 129 QNALNGRFFGGRQIHAEFATDAMINAVCGNSSTNT 163
NAL+GR+F G+ I A + + + +S T T
Sbjct 488 VNALHGRWFAGKMITAAYVPLPTYHNLFPDSMTAT 522
> pfa:MAL13P1.120 splicing factor, putative; K13091 RNA-binding
protein 39
Length=864
Score = 65.1 bits (157), Expect = 1e-10, Method: Composition-based stats.
Identities = 34/93 (36%), Positives = 57/93 (61%), Gaps = 3/93 (3%)
Query 67 LVLRGMF---NPSAADDEYYFEDIRDDVREECGKHGSVVQVSLSEKDEEGRVFVKFTAPS 123
LVL MF + + D +F DI +DV+EEC K+G VV + L K+ +G++++K++
Sbjct 769 LVLSNMFSSNDENIGSDPDFFNDILEDVKEECSKYGKVVNIWLDTKNIDGKIYIKYSNND 828
Query 124 VALAAQNALNGRFFGGRQIHAEFATDAMINAVC 156
+L + LNGR+FGG I+A F ++ + + C
Sbjct 829 ESLKSFQFLNGRYFGGSLINAYFISNDVWDMTC 861
Score = 30.4 bits (67), Expect = 3.3, Method: Composition-based stats.
Identities = 16/51 (31%), Positives = 25/51 (49%), Gaps = 0/51 (0%)
Query 112 EGRVFVKFTAPSVALAAQNALNGRFFGGRQIHAEFATDAMINAVCGNSSTN 162
+G F++F S A+ A +NG GR+I +A D+ C N+ N
Sbjct 630 KGFGFIQFHKASEAIEALTVMNGMEVAGREIKVGYAQDSKYLLACDNTQEN 680
> tgo:TGME49_002770 RNA-binding protein, putative ; K12838 poly(U)-binding-splicing
factor PUF60
Length=532
Score = 64.7 bits (156), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 37/86 (43%), Positives = 52/86 (60%), Gaps = 10/86 (11%)
Query 59 APGVMTSNLVLRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSLSEKDEEGRVFVK 118
+P V+ SN+V PS D E ++D+VREEC K GS+ +V + E R+FV+
Sbjct 442 SPVVLLSNMV-----TPSEVDGE-----LKDEVREECSKFGSIKRVEVHTLKETVRIFVE 491
Query 119 FTAPSVALAAQNALNGRFFGGRQIHA 144
F+ S A A +L+GR+FGGRQI A
Sbjct 492 FSDLSGAREAIPSLHGRWFGGRQIIA 517
> dre:394178 htatsf1, MGC66252, zgc:66252; HIV TAT specific factor
1; K13093 HIV Tat-specific factor 1
Length=450
Score = 63.9 bits (154), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 31/79 (39%), Positives = 48/79 (60%), Gaps = 1/79 (1%)
Query 67 LVLRGMFNPSA-ADDEYYFEDIRDDVREECGKHGSVVQVSLSEKDEEGRVFVKFTAPSVA 125
++++ MF+P+ +D + RDD+R EC K G V +V + ++ +G V F P A
Sbjct 270 IIIQNMFHPTDFEEDPLVLNEYRDDLRTECEKFGQVKKVIIFDRHPDGVASVAFKEPEEA 329
Query 126 LAAQNALNGRFFGGRQIHA 144
A Q ALNGR+FGGR++ A
Sbjct 330 DACQVALNGRWFGGRKLSA 348
> ath:AT1G30480 DRT111; DRT111; nucleic acid binding / nucleotide
binding; K12840 splicing factor 45
Length=387
Score = 63.9 bits (154), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 36/93 (38%), Positives = 49/93 (52%), Gaps = 12/93 (12%)
Query 61 GVMTSNLVLRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSLSEKDEEG------- 113
G T L+LR M P DDE + D+V ECGK+G+V +V + E E
Sbjct 279 GEPTRVLLLRNMVGPGQVDDE-----LEDEVGGECGKYGTVTRVLIFEITEPNFPVHEAV 333
Query 114 RVFVKFTAPSVALAAQNALNGRFFGGRQIHAEF 146
R+FV+F+ P A L+GR+FGGR + A F
Sbjct 334 RIFVQFSRPEETTKALVDLDGRYFGGRTVRATF 366
> mmu:67959 Puf60, 2410104I19Rik, 2810454F19Rik, SIAHBP1; poly-U
binding splicing factor 60; K12838 poly(U)-binding-splicing
factor PUF60
Length=499
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 36/87 (41%), Positives = 51/87 (58%), Gaps = 13/87 (14%)
Query 67 LVLRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSL------SEKDEE--GRVFVK 118
+VLR M +P DD D+ +V EECGK G+V +V + E+D E ++FV+
Sbjct 404 MVLRNMVDPKDIDD-----DLEGEVTEECGKFGAVNRVIIYQEKQGEEEDAEIIVKIFVE 458
Query 119 FTAPSVALAAQNALNGRFFGGRQIHAE 145
F+ S A ALNGR+FGGR++ AE
Sbjct 459 FSMASETHKAIQALNGRWFGGRKVVAE 485
Score = 31.6 bits (70), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 17/64 (26%), Positives = 27/64 (42%), Gaps = 4/64 (6%)
Query 83 YFEDIRDDVREECGKHGSVVQVSLSE----KDEEGRVFVKFTAPSVALAAQNALNGRFFG 138
Y+E D +R+ G + + +S +G FV++ P A A +N G
Sbjct 77 YYELGEDTIRQAFAPFGPIKSIDMSWDSVTMKHKGFAFVEYEVPEAAQLALEQMNSVMLG 136
Query 139 GRQI 142
GR I
Sbjct 137 GRNI 140
> dre:562370 puf60a, fc21a05, wu:fc21a05, wu:fi43a08; poly-U binding
splicing factor a; K12838 poly(U)-binding-splicing factor
PUF60
Length=518
Score = 59.7 bits (143), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 44/132 (33%), Positives = 67/132 (50%), Gaps = 29/132 (21%)
Query 26 QQQSQQQQHVQASPSTDK----QQFLRSSSGPRVGGLAPGVMTSNLVLRGMFNPSAADDE 81
Q+ +Q+H+ S S+ + Q+ LR ++ +VLR M P DD
Sbjct 390 QEMLSEQEHMSISGSSARHMVMQKLLRKQE------------STVMVLRNMVGPEDIDD- 436
Query 82 YYFEDIRDDVREECGKHGSVVQVSL------SEKDEEG--RVFVKFTAPSVALAAQNALN 133
D+ +V EECGK G+V +V + E+D E ++FV+F+A S A ALN
Sbjct 437 ----DLEGEVTEECGKFGAVNRVIIYQEKQGEEEDAEVIVKIFVEFSAASEMNKAIQALN 492
Query 134 GRFFGGRQIHAE 145
R+FGGR++ AE
Sbjct 493 NRWFGGRKVIAE 504
Score = 31.6 bits (70), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 17/64 (26%), Positives = 27/64 (42%), Gaps = 4/64 (6%)
Query 83 YFEDIRDDVREECGKHGSVVQVSLSEK----DEEGRVFVKFTAPSVALAAQNALNGRFFG 138
Y+E D +R+ G + + +S +G FV++ P A A +N G
Sbjct 100 YYELGEDTIRQAFAPFGPIKSIDMSWDSVTLKHKGFAFVEYEVPEAAQLALEQMNSVMLG 159
Query 139 GRQI 142
GR I
Sbjct 160 GRNI 163
> cpv:cgd1_2730 Ro ribonucleoprotein-binding protein 1, RNA binding
protein with 3x RRM domains ; K12838 poly(U)-binding-splicing
factor PUF60
Length=693
Score = 59.7 bits (143), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 31/85 (36%), Positives = 49/85 (57%), Gaps = 11/85 (12%)
Query 64 TSNLVLRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQV------SLSEKDEEGRVFV 117
T+ ++L M P DDE ++++V+ EC K+G V V ++S+ + R+FV
Sbjct 597 TNIILLTNMVGPDEIDDE-----LKEEVKIECSKYGKVYDVRIHVSNNISKPSDRVRIFV 651
Query 118 KFTAPSVALAAQNALNGRFFGGRQI 142
F +PS+A A ALN R+FGG Q+
Sbjct 652 VFESPSMAQIAVPALNNRWFGGNQV 676
> hsa:22827 PUF60, FIR, FLJ31379, RoBPI, SIAHBP1; poly-U binding
splicing factor 60KDa; K12838 poly(U)-binding-splicing factor
PUF60
Length=516
Score = 59.3 bits (142), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 35/87 (40%), Positives = 50/87 (57%), Gaps = 13/87 (14%)
Query 67 LVLRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSL------SEKDEE--GRVFVK 118
+VLR M +P DD D+ +V EECGK G+V +V + E+D E ++FV+
Sbjct 421 MVLRNMVDPKDIDD-----DLEGEVTEECGKFGAVNRVIIYQEKQGEEEDAEIIVKIFVE 475
Query 119 FTAPSVALAAQNALNGRFFGGRQIHAE 145
F+ S A ALNGR+F GR++ AE
Sbjct 476 FSIASETHKAIQALNGRWFAGRKVVAE 502
Score = 31.6 bits (70), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 17/64 (26%), Positives = 27/64 (42%), Gaps = 4/64 (6%)
Query 83 YFEDIRDDVREECGKHGSVVQVSLSE----KDEEGRVFVKFTAPSVALAAQNALNGRFFG 138
Y+E D +R+ G + + +S +G FV++ P A A +N G
Sbjct 94 YYELGEDTIRQAFAPFGPIKSIDMSWDSVTMKHKGFAFVEYEVPEAAQLALEQMNSVMLG 153
Query 139 GRQI 142
GR I
Sbjct 154 GRNI 157
> cpv:cgd7_5220 splicing factor with 3 RRM domains ; K13091 RNA-binding
protein 39
Length=563
Score = 59.3 bits (142), Expect = 6e-09, Method: Composition-based stats.
Identities = 31/91 (34%), Positives = 53/91 (58%), Gaps = 3/91 (3%)
Query 62 VMTSNLVLRGMFNPSAADDE---YYFEDIRDDVREECGKHGSVVQVSLSEKDEEGRVFVK 118
++ SN+ S +DE E+I+ DV EECGK+G++++ L ++ +G V+VK
Sbjct 464 LLLSNMFTEQSIKESMEEDETIEQILEEIQADVEEECGKYGTLLECFLDKEKMDGNVWVK 523
Query 119 FTAPSVALAAQNALNGRFFGGRQIHAEFATD 149
++ P A A+ +GRFF GR+++ F D
Sbjct 524 YSRPEEASKAKMVFHGRFFAGRKLNVSFIKD 554
> cel:Y47G6A.20 rnp-6; RNP (RRM RNA binding domain) containing
family member (rnp-6); K12838 poly(U)-binding-splicing factor
PUF60
Length=749
Score = 58.2 bits (139), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 31/81 (38%), Positives = 47/81 (58%), Gaps = 7/81 (8%)
Query 67 LVLRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSLSEKDEEG--RVFVKFTAPSV 124
+VLR M P D E + ++REECGK+G+V+ V ++ G ++FVK++
Sbjct 660 IVLRNMVTPQDID-----EFLEGEIREECGKYGNVIDVVIANFASSGLVKIFVKYSDSMQ 714
Query 125 ALAAQNALNGRFFGGRQIHAE 145
A+ AL+GRFFGG + AE
Sbjct 715 VDRAKAALDGRFFGGNTVKAE 735
Score = 36.6 bits (83), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 19/65 (29%), Positives = 30/65 (46%), Gaps = 4/65 (6%)
Query 84 FEDIRDDVREECGKHGSVVQVSLSEKDEEGR----VFVKFTAPSVALAAQNALNGRFFGG 139
FE D +R G + +++S G FV++ P AL AQ ++NG+ GG
Sbjct 111 FEIREDMLRRAFDPFGPIKSINMSWDPATGHHKTFAFVEYEVPEAALLAQESMNGQMLGG 170
Query 140 RQIHA 144
R +
Sbjct 171 RNLKV 175
> tpv:TP04_0540 hypothetical protein
Length=486
Score = 56.2 bits (134), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 38/91 (41%), Positives = 51/91 (56%), Gaps = 11/91 (12%)
Query 58 LAPG---VMTSN-LVLRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSL--SEKDE 111
LAP V TSN +V+ M +P AD E++ ++V+ EC K+G+V V L S ++
Sbjct 384 LAPESSVVGTSNVIVIYNMVDPKLAD-----ENLANEVKVECNKYGTVTSVYLHFSANND 438
Query 112 EGRVFVKFTAPSVALAAQNALNGRFFGGRQI 142
VFV F P A A ALN R+F GRQI
Sbjct 439 TLSVFVVFNTPEDADNAVRALNTRWFNGRQI 469
> mmu:16589 Uhmk1, 4732477C12Rik, 4930500M09Rik, AA673513, AI449218,
AU021979, KIS, Kist; U2AF homology motif (UHM) kinase
1 (EC:2.7.11.1); K08877 U2AF homology motif (UHM) kinase 1
[EC:2.7.11.1]
Length=419
Score = 55.8 bits (133), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 39/108 (36%), Positives = 55/108 (50%), Gaps = 14/108 (12%)
Query 46 FLRSSSGPRVGGLAPGVMTSNLVLRGMFNPSAADDEYY-----FEDIRDDVREECGKHGS 100
F P + L VM VLR + + DD+Y +ED+ +DV+EEC K+G
Sbjct 303 FFSIPFAPHIEDL---VMLPTPVLRLL---NVLDDDYLENEDEYEDVVEDVKEECQKYGP 356
Query 101 VVQVSLSEKDEEGR--VFVKFTAPSVALAAQNALNGRFFGGRQIHAEF 146
VV + L K+ GR VFV++ + AAQ L GR F G+ + A F
Sbjct 357 VVSL-LVPKENPGRGQVFVEYANAGDSKAAQKLLTGRMFDGKFVVATF 403
> ath:AT5G09880 RNA recognition motif (RRM)-containing protein;
K13091 RNA-binding protein 39
Length=527
Score = 55.5 bits (132), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 30/86 (34%), Positives = 53/86 (61%), Gaps = 1/86 (1%)
Query 61 GVMTSNLVLRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSLSEKDEEGRVFVKFT 120
G+ + L+L+ MF+P+ + + +IRDDV +EC K+G V + + +K+ G V+++F
Sbjct 431 GLPSECLLLKNMFDPATETEPNFDLEIRDDVADECSKYGPVNHIYV-DKNSAGFVYLRFQ 489
Query 121 APSVALAAQNALNGRFFGGRQIHAEF 146
+ A AAQ A++ R+F + I A F
Sbjct 490 SVEAAAAAQRAMHMRWFAQKMISATF 515
> dre:393889 rbm17, MGC55840, zgc:55840; RNA binding motif protein
17; K12840 splicing factor 45
Length=418
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 32/88 (36%), Positives = 46/88 (52%), Gaps = 10/88 (11%)
Query 64 TSNLVLRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSLSE-----KDEEGRVFVK 118
T ++LR M D ED+ + +EEC K+G VV+ + E DE R+F++
Sbjct 321 TKVVLLRNMVGRGEVD-----EDLEAETKEECEKYGKVVRCVIFEISGVTDDEAVRIFLE 375
Query 119 FTAPSVALAAQNALNGRFFGGRQIHAEF 146
F A+ A LNGR+FGGR + A F
Sbjct 376 FERVESAIKAVVDLNGRYFGGRVVKACF 403
> xla:398736 htatsf1, MGC98898; HIV-1 Tat specific factor 1; K13093
HIV Tat-specific factor 1
Length=452
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 27/80 (33%), Positives = 45/80 (56%), Gaps = 1/80 (1%)
Query 67 LVLRGMFNPSA-ADDEYYFEDIRDDVREECGKHGSVVQVSLSEKDEEGRVFVKFTAPSVA 125
++++ MF+P +D +IR+D+R EC K G V ++ + ++ +G V F +
Sbjct 271 VIIKNMFHPKDFEEDPLVLNEIREDLRSECEKFGQVKKLLIFDQHPDGVASVAFKEANEG 330
Query 126 LAAQNALNGRFFGGRQIHAE 145
ALNGR+FGGRQ+ E
Sbjct 331 DLCIQALNGRWFGGRQLAVE 350
> dre:415211 puf60b, SIAHBP1, fe37c05, repressor, si:ch211-12p8.2,
si:zc12p8.2, wu:fb33e11, wu:fe37c05, zgc:86806; poly-U
binding splicing factor b
Length=502
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 33/88 (37%), Positives = 47/88 (53%), Gaps = 13/88 (14%)
Query 67 LVLRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSL------SEKDEE--GRVFVK 118
+VLR M P DD D+ +V EECGK+G+V +V + E D E ++FV+
Sbjct 407 MVLRNMVGPEDIDD-----DLEGEVMEECGKYGAVNRVIIYQERQGEEDDAEIIVKIFVE 461
Query 119 FTAPSVALAAQNALNGRFFGGRQIHAEF 146
F+ A ALN R+F GR++ AE
Sbjct 462 FSDAGEMNKAIQALNNRWFAGRKVVAEL 489
Score = 31.2 bits (69), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 17/64 (26%), Positives = 27/64 (42%), Gaps = 4/64 (6%)
Query 83 YFEDIRDDVREECGKHGSVVQVSLSE----KDEEGRVFVKFTAPSVALAAQNALNGRFFG 138
Y+E D +R+ G + + +S +G FV++ P A A +N G
Sbjct 77 YYELGEDTIRQAFAPFGPIKSIDMSWDSVTMKHKGFAFVEYEVPEAAQLALEQMNSVMLG 136
Query 139 GRQI 142
GR I
Sbjct 137 GRNI 140
> mmu:72459 Htatsf1, 1600023H17Rik, 2600017A12Rik, 2700077B20Rik,
TAT-SF1; HIV TAT specific factor 1; K13093 HIV Tat-specific
factor 1
Length=757
Score = 53.9 bits (128), Expect = 3e-07, Method: Composition-based stats.
Identities = 30/80 (37%), Positives = 46/80 (57%), Gaps = 1/80 (1%)
Query 67 LVLRGMFNP-SAADDEYYFEDIRDDVREECGKHGSVVQVSLSEKDEEGRVFVKFTAPSVA 125
++L+ MF+P DD +IR+D+R EC K G + ++ L ++ +G V F P A
Sbjct 267 VILKNMFHPMDFEDDPLVLNEIREDLRVECSKFGQIRKLLLFDRHPDGVASVSFREPEEA 326
Query 126 LAAQNALNGRFFGGRQIHAE 145
L+GR+FGGRQI A+
Sbjct 327 DHCIQTLDGRWFGGRQITAQ 346
> mmu:380664 Lemd3, AI316861, Man1; LEM domain containing 3
Length=918
Score = 53.9 bits (128), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 24/87 (27%), Positives = 47/87 (54%), Gaps = 0/87 (0%)
Query 60 PGVMTSNLVLRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSLSEKDEEGRVFVKF 119
P +T L +R MF+P +++ I++ + E+C + +V +++ EG V+VK
Sbjct 786 PNSLTPCLKIRNMFDPVMEIGDHWHLAIQEAILEKCSDNDGIVHIAVDRNSREGCVYVKC 845
Query 120 TAPSVALAAQNALNGRFFGGRQIHAEF 146
+P A A AL+G +F G+ + ++
Sbjct 846 LSPEYAGKAFKALHGSWFDGKLVTVKY 872
> hsa:84991 RBM17, DKFZp686F13131, MGC14439, SPF45; RNA binding
motif protein 17; K12840 splicing factor 45
Length=401
Score = 53.1 bits (126), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 41/132 (31%), Positives = 63/132 (47%), Gaps = 12/132 (9%)
Query 20 STASDSQQQSQQQQHVQASPSTDKQQFLRSSSGPRVGGLAPGVMTSNLVLRGMFNPSAAD 79
STA ++ S++ + +T+K +S S P L T ++LR M D
Sbjct 262 STALSVEKTSKRGGKIIVGDATEKDASKKSDSNPLTEILK--CPTKVVLLRNMVGAGEVD 319
Query 80 DEYYFEDIRDDVREECGKHGSVVQVSLSE-----KDEEGRVFVKFTAPSVALAAQNALNG 134
ED+ + +EEC K+G V + + E DE R+F++F A+ A LNG
Sbjct 320 -----EDLEVETKEECEKYGKVGKCVIFEIPGAPDDEAVRIFLEFERVESAIKAVVDLNG 374
Query 135 RFFGGRQIHAEF 146
R+FGGR + A F
Sbjct 375 RYFGGRVVKACF 386
> hsa:23592 LEMD3, MAN1; LEM domain containing 3
Length=910
Score = 53.1 bits (126), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 24/87 (27%), Positives = 47/87 (54%), Gaps = 0/87 (0%)
Query 60 PGVMTSNLVLRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSLSEKDEEGRVFVKF 119
P +T L +R MF+P + + I++ + E+C + +V +++ + EG V+VK
Sbjct 778 PNSLTPCLKIRNMFDPVMEIGDQWHLAIQEAILEKCSDNDGIVHIAVDKNSREGCVYVKC 837
Query 120 TAPSVALAAQNALNGRFFGGRQIHAEF 146
+P A A AL+G +F G+ + ++
Sbjct 838 LSPEYAGKAFKALHGSWFDGKLVTVKY 864
> ath:AT5G16260 RNA recognition motif (RRM)-containing protein;
K13093 HIV Tat-specific factor 1
Length=519
Score = 52.8 bits (125), Expect = 6e-07, Method: Composition-based stats.
Identities = 31/85 (36%), Positives = 46/85 (54%), Gaps = 1/85 (1%)
Query 63 MTSNLVLRGMFNPSA-ADDEYYFEDIRDDVREECGKHGSVVQVSLSEKDEEGRVFVKFTA 121
+ + +VLR MF+P+ DE ++ +DV+EE KHG V + E +G V V+F
Sbjct 404 IPATVVLRYMFSPAELMADEDLVAELEEDVKEESLKHGPFDSVKVCEHHPQGVVLVRFKD 463
Query 122 PSVALAAQNALNGRFFGGRQIHAEF 146
A A+NGR++ RQIHA
Sbjct 464 RRDAQKCIEAMNGRWYAKRQIHASL 488
> dre:402989 u2af2b, MGC77804, wu:fb73g02, zgc:77804; U2 small
nuclear RNA auxiliary factor 2b; K12837 splicing factor U2AF
65 kDa subunit
Length=475
Score = 52.8 bits (125), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 28/93 (30%), Positives = 46/93 (49%), Gaps = 6/93 (6%)
Query 61 GVMTSNLVLRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSLSEKDE------EGR 114
G+ T L L M P D+ +E+I +DV+EEC K+G V + + + G+
Sbjct 372 GIPTEVLCLMNMVAPEELIDDEEYEEIVEDVKEECSKYGQVKSIEIPRPVDGLDIPGTGK 431
Query 115 VFVKFTAPSVALAAQNALNGRFFGGRQIHAEFA 147
+FV+FT+ + A L GR F R + ++
Sbjct 432 IFVEFTSVYDSQKAMQGLTGRKFANRVVVTKYC 464
> hsa:27336 HTATSF1, TAT-SF1, dJ196E23.2; HIV-1 Tat specific factor
1; K13093 HIV Tat-specific factor 1
Length=755
Score = 52.4 bits (124), Expect = 8e-07, Method: Composition-based stats.
Identities = 29/80 (36%), Positives = 46/80 (57%), Gaps = 1/80 (1%)
Query 67 LVLRGMFNP-SAADDEYYFEDIRDDVREECGKHGSVVQVSLSEKDEEGRVFVKFTAPSVA 125
++++ MF+P DD +IR+D+R EC K G + ++ L ++ +G V F P A
Sbjct 266 VIIKNMFHPMDFEDDPLVLNEIREDLRVECSKFGQIRKLLLFDRHPDGVASVSFRDPEEA 325
Query 126 LAAQNALNGRFFGGRQIHAE 145
L+GR+FGGRQI A+
Sbjct 326 DYCIQTLDGRWFGGRQITAQ 345
> xla:380287 u2af2, MGC53441, u2af65; U2 small nuclear RNA auxiliary
factor 2; K12837 splicing factor U2AF 65 kDa subunit
Length=456
Score = 52.0 bits (123), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 32/105 (30%), Positives = 50/105 (47%), Gaps = 16/105 (15%)
Query 59 APGVMTSN----------LVLRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSLS- 107
PG+M+S L L M P D+ +E+I +DVR+ECGK+G+V + +
Sbjct 341 VPGLMSSQVQMGGHPTEVLCLMNMVVPEELIDDDEYEEIVEDVRDECGKYGAVKSIEIPR 400
Query 108 -----EKDEEGRVFVKFTAPSVALAAQNALNGRFFGGRQIHAEFA 147
E G++FV+FT+ A L GR F R + ++
Sbjct 401 PVDGVEVPGCGKIFVEFTSVFDCQKAMQGLTGRKFANRVVVTKYC 445
> cel:F58B3.7 hypothetical protein; K12840 splicing factor 45
Length=371
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 28/63 (44%), Positives = 39/63 (61%), Gaps = 5/63 (7%)
Query 89 DDVREECGKHGSVVQVSL-----SEKDEEGRVFVKFTAPSVALAAQNALNGRFFGGRQIH 143
D+++EE K G VV V + E+D + RVFV+FT + A+ A +NGRFFGGR +
Sbjct 298 DEIKEEMEKCGQVVNVIVHVDESQEEDRQVRVFVEFTNNAQAIKAFVMMNGRFFGGRSVS 357
Query 144 AEF 146
A F
Sbjct 358 AGF 360
> cel:Y92C3B.2 uaf-1; U2AF splicing factor family member (uaf-1);
K12837 splicing factor U2AF 65 kDa subunit
Length=474
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 30/71 (42%), Positives = 41/71 (57%), Gaps = 8/71 (11%)
Query 78 ADDEYYFEDIRDDVREECGKHGSVVQVSLSEKDEE------GRVFVKFTAPSVALAAQNA 131
ADDEY E+I +DVR+EC K+G V + + E+ G+VFV+F + S AQ A
Sbjct 391 ADDEY--EEILEDVRDECSKYGIVRSLEIPRPYEDHPVPGVGKVFVEFASTSDCQRAQAA 448
Query 132 LNGRFFGGRQI 142
L GR F R +
Sbjct 449 LTGRKFANRTV 459
> bbo:BBOV_III001520 17.m07155; splicing factor, CC1-like family
protein; K13091 RNA-binding protein 39
Length=488
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 33/88 (37%), Positives = 53/88 (60%), Gaps = 2/88 (2%)
Query 64 TSNLVLRGMFNPS--AADDEYYFEDIRDDVREECGKHGSVVQVSLSEKDEEGRVFVKFTA 121
T N+ L MF+ S + + +F+++ +DV EEC K+G V++V ++ +G+V+VKF
Sbjct 393 TCNITLSNMFSSSDPSVSEPTFFDEVEEDVNEECNKYGKVLKVYINRGVIDGKVWVKFGN 452
Query 122 PSVALAAQNALNGRFFGGRQIHAEFATD 149
+ A +LNGR F G I AE+ TD
Sbjct 453 VVDSTVAFRSLNGRVFAGNTIKAEYVTD 480
Score = 28.9 bits (63), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 18/65 (27%), Positives = 31/65 (47%), Gaps = 4/65 (6%)
Query 90 DVREECGKHGSVVQVSLSEKDEE----GRVFVKFTAPSVALAAQNALNGRFFGGRQIHAE 145
++R+ G+++ V + G+ ++KF S A A A+NG GG+ I
Sbjct 263 EIRQMFSPFGNIISVEILRDPHSNLPLGQAYIKFKRTSEAKEAVTAMNGFDIGGQTIKVA 322
Query 146 FATDA 150
+AT A
Sbjct 323 YATGA 327
> mmu:76938 Rbm17, 2700027J02Rik; RNA binding motif protein 17;
K12840 splicing factor 45
Length=405
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 31/88 (35%), Positives = 45/88 (51%), Gaps = 10/88 (11%)
Query 64 TSNLVLRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSLSE-----KDEEGRVFVK 118
T ++LR M D ED+ + +EEC K+G V + + E DE R+F++
Sbjct 308 TKVVLLRNMVGAGEVD-----EDLEVETKEECEKYGKVGKCVIFEIPGAPDDEAVRIFLE 362
Query 119 FTAPSVALAAQNALNGRFFGGRQIHAEF 146
F A+ A LNGR+FGGR + A F
Sbjct 363 FERVESAIKAVVDLNGRYFGGRVVKACF 390
> tpv:TP03_0201 hypothetical protein
Length=509
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 37/122 (30%), Positives = 55/122 (45%), Gaps = 15/122 (12%)
Query 62 VMTSNLVLRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSLSE-----KDEE---- 112
+ T L+L + + +D+ + DI DDVR EC +G V++V L +EE
Sbjct 357 IPTRVLLLSNLVSKDELEDDEEYVDIIDDVRCECELYGVVLRVELPRVPKGLTEEEMKAF 416
Query 113 -----GRVFVKFTAPSVALAAQNALNGRFFGGRQIHAEFATDA-MINAVCGNSSTNTGNN 166
G FV F+ A A+ L+GR FG R +HA F ++ + GN N
Sbjct 417 DPTSVGSAFVLFSTVESASKARKVLDGRKFGQRTVHAHFFSELYFLTGKFGNPKPNFAKE 476
Query 167 SS 168
S
Sbjct 477 HS 478
> dre:767721 uhmk1, MGC153241, zgc:153241; U2AF homology motif
(UHM) kinase 1 (EC:2.7.11.1); K08877 U2AF homology motif (UHM)
kinase 1 [EC:2.7.11.1]
Length=410
Score = 50.4 bits (119), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 27/69 (39%), Positives = 45/69 (65%), Gaps = 3/69 (4%)
Query 79 DDEYYFEDIRDDVREECGKHGSVVQVSLSEKDE-EGRVFVKFTAPSVALAAQNALNGRFF 137
+DEY EDI +D++EEC K+G+VV + + +++ +G+VFV++ + AQ L GR F
Sbjct 328 EDEY--EDIIEDMKEECQKYGTVVSLLIPKENPGKGQVFVEYANAGDSKEAQRLLTGRTF 385
Query 138 GGRQIHAEF 146
G+ + A F
Sbjct 386 DGKFVVATF 394
> tgo:TGME49_121630 hypothetical protein
Length=400
Score = 48.9 bits (115), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 29/92 (31%), Positives = 45/92 (48%), Gaps = 10/92 (10%)
Query 57 GLAPGVMTSNLVLRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSLS--EKDEEGR 114
G AP + +VL+ + + DD D++D++ +EC KHG VV+V + +E R
Sbjct 304 GDAPSTL---IVLKNLMEVAELDD-----DVKDEIYQECLKHGKVVEVRIHVVASTQEVR 355
Query 115 VFVKFTAPSVALAAQNALNGRFFGGRQIHAEF 146
F + P A A LN R F R++ E
Sbjct 356 AFALYQLPEQANRAVRVLNERSFAKRKVKCEL 387
> tgo:TGME49_036910 U2 snRNP auxiliary factor small subunit, putative
; K12836 splicing factor U2AF 35 kDa subunit
Length=254
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 36/102 (35%), Positives = 50/102 (49%), Gaps = 19/102 (18%)
Query 64 TSNLVLRGMF-NPSAA---------DDEY------YFEDIRDDVREECGKHGSVVQVSLS 107
+ +VLR M+ NP A DE +FE +V EE K+G V + +
Sbjct 43 SPTIVLRHMYPNPPVAVAIAEGQNVSDELLDQAADHFEAFFSEVFEELAKYGEVEDMVVC 102
Query 108 EKDEE---GRVFVKFTAPSVALAAQNALNGRFFGGRQIHAEF 146
+ + G V+VK+T A A AL GRF+ G+QIHAEF
Sbjct 103 DNIGDHIIGNVYVKYTDEEAANKALAALQGRFYSGKQIHAEF 144
> xla:398582 lemd3, man1, sane, xman1; LEM domain containing 3
Length=781
Score = 47.8 bits (112), Expect = 2e-05, Method: Composition-based stats.
Identities = 24/87 (27%), Positives = 48/87 (55%), Gaps = 0/87 (0%)
Query 60 PGVMTSNLVLRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSLSEKDEEGRVFVKF 119
P +T L +R MF+P +++ I++ + E+C + +V +++ + EG V+VK
Sbjct 649 PNSLTPCLKIRNMFDPVMEIGDHWDLAIQEAILEKCSDNEGIVHIAVDKNSREGCVYVKC 708
Query 120 TAPSVALAAQNALNGRFFGGRQIHAEF 146
+P A A AL+G +F G+ + ++
Sbjct 709 LSPEFAGKAFKALHGSWFDGKLVTVKY 735
> xla:779027 sane, LEMD3; Smad1 antagonistic effector
Length=784
Score = 47.8 bits (112), Expect = 2e-05, Method: Composition-based stats.
Identities = 24/87 (27%), Positives = 48/87 (55%), Gaps = 0/87 (0%)
Query 60 PGVMTSNLVLRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSLSEKDEEGRVFVKF 119
P +T L +R MF+P +++ I++ + E+C + +V +++ + EG V+VK
Sbjct 652 PNSLTPCLKIRNMFDPVMEIGDHWDLAIQEAILEKCSDNEGIVHIAVDKNSREGCVYVKC 711
Query 120 TAPSVALAAQNALNGRFFGGRQIHAEF 146
+P A A AL+G +F G+ + ++
Sbjct 712 LSPEFAGKAFKALHGSWFDGKLVTVKY 738
> dre:558474 lemd3, im:7158446, si:dkey-6a5.6, wu:fb34b12; LEM
domain containing 3
Length=841
Score = 47.4 bits (111), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 23/87 (26%), Positives = 46/87 (52%), Gaps = 0/87 (0%)
Query 60 PGVMTSNLVLRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSLSEKDEEGRVFVKF 119
P +T L +R MF+P E + I++ + E+C + +V +++ + EG V+VK
Sbjct 708 PNSLTPCLKIRNMFDPVMEVGENWHLAIQEAILEKCSDNDGIVHIAVDKNSREGCVYVKC 767
Query 120 TAPSVALAAQNALNGRFFGGRQIHAEF 146
+ + A AL+G +F G+ + ++
Sbjct 768 LSAEHSGKAFKALHGSWFDGKLVTVKY 794
> hsa:8233 ZRSR2, MGC142014, MGC142040, U2AF1-RS2, U2AF1L2, U2AF1RS2,
URP; zinc finger (CCCH type), RNA-binding motif and
serine/arginine rich 2
Length=482
Score = 46.6 bits (109), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 31/98 (31%), Positives = 48/98 (48%), Gaps = 10/98 (10%)
Query 70 RGMFNPSAA----DDEYY--FEDIRDDVREECGKHGSVVQVSLS---EKDEEGRVFVKFT 120
R ++P A+ ++E Y F D +DV E G V+Q +S E G V+V++
Sbjct 216 RDDYDPDASLEYSEEETYQQFLDFYEDVLPEFKNVGKVIQFKVSCNLEPHLRGNVYVQYQ 275
Query 121 APSVALAAQNALNGRFFGGRQIHAEFATDAMIN-AVCG 157
+ AA + NGR++ GRQ+ EF A+CG
Sbjct 276 SEEECQAALSLFNGRWYAGRQLQCEFCPVTRWKMAICG 313
> ath:AT1G60900 U2 snRNP auxiliary factor large subunit, putative;
K12837 splicing factor U2AF 65 kDa subunit
Length=589
Score = 45.4 bits (106), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 26/75 (34%), Positives = 44/75 (58%), Gaps = 8/75 (10%)
Query 79 DDEYYFEDIRDDVREECGKHGSVVQVSLSEKDEE-------GRVFVKFTAPSVALAAQNA 131
DDE Y E I +D+R+E GK G++V V + + + G+VF+++ + A++
Sbjct 503 DDEEYAE-IMEDMRQEGGKFGNLVNVVIPRPNPDHDPTPGVGKVFLEYADVDGSSKARSG 561
Query 132 LNGRFFGGRQIHAEF 146
+NGR FGG Q+ A +
Sbjct 562 MNGRKFGGNQVVAVY 576
> bbo:BBOV_IV007070 23.m05938; hypothetical protein
Length=400
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 31/107 (28%), Positives = 50/107 (46%), Gaps = 14/107 (13%)
Query 62 VMTSNLVLRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSLSE------KDE---- 111
+ T L+L + + +D+ + DI DDVR EC ++G VV+V + DE
Sbjct 253 IPTRVLLLANLVSKEDLEDDAEYYDIIDDVRCECEEYGPVVRVEMPRVPKGLTLDEIRNM 312
Query 112 ----EGRVFVKFTAPSVALAAQNALNGRFFGGRQIHAEFATDAMINA 154
G FV F+ A A+ L+GR FG R + F ++ + +
Sbjct 313 DFSAVGCAFVLFSNIEGASKARKVLDGRKFGHRIVECHFFSELLFHV 359
> pfa:PF11_0200 U2 snRNP auxiliary factor, small subunit, putative;
K12836 splicing factor U2AF 35 kDa subunit
Length=294
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 29/97 (29%), Positives = 49/97 (50%), Gaps = 11/97 (11%)
Query 53 PRVGGLAPGVMTSNLVLRGMFNPSAADDEYYFEDIRDDVREECGKHGSVVQVSLSEKDEE 112
P +A G M + VL AAD +FE+ ++V +E K+G + + + + +
Sbjct 56 PIAVAIAEGQMVEDEVL-----DKAAD---HFEEFYEEVFDELMKYGEIEDMVVCDNIGD 107
Query 113 ---GRVFVKFTAPSVALAAQNALNGRFFGGRQIHAEF 146
G V++K+T A A N LNGRF+ G+ + E+
Sbjct 108 HIIGNVYIKYTHEDYAEKAVNELNGRFYAGKPLQIEY 144
> mmu:22183 Zrsr1, D11Ncvs75, Irlgs2, SP2, U2af1-rs1, U2afbp-rs;
zinc finger (CCCH type), RNA binding motif and serine/arginine
rich 1
Length=428
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 43/90 (47%), Gaps = 6/90 (6%)
Query 74 NPSAADDEYY--FEDIRDDVREECGKHGSVVQVSLS---EKDEEGRVFVKFTAPSVALAA 128
N +++E Y F D DV E G V+Q +S E G V+V++ + AA
Sbjct 215 NLEYSEEETYQQFLDFYHDVLPEFKNVGKVIQFKVSCNLEPHLRGNVYVQYQSEEECQAA 274
Query 129 QNALNGRFFGGRQIHAEFATDAMIN-AVCG 157
+ NGR++ GRQ+ EF A+CG
Sbjct 275 LSLFNGRWYAGRQLQCEFCPVTRWKVAICG 304
Lambda K H
0.310 0.123 0.337
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4341553636
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40