bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0130_orf4
Length=167
Score E
Sequences producing significant alignments: (Bits) Value
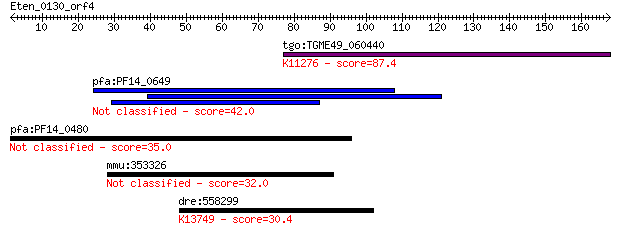
tgo:TGME49_060440 46 kDa FK506-binding nuclear protein, putati... 87.4 2e-17
pfa:PF14_0649 conserved Plasmodium protein, unknown function 42.0 0.001
pfa:PF14_0480 conserved Plasmodium protein, unknown function 35.0 0.12
mmu:353326 Rtl1, 6430411K18Rik, Mar, Mart1, Mor1, Peg11; retro... 32.0 0.93
dre:558299 slc24a1, si:ch211-117i10.3; solute carrier family 2... 30.4 2.9
> tgo:TGME49_060440 46 kDa FK506-binding nuclear protein, putative
(EC:5.2.1.8); K11276 nucleophosmin 1
Length=311
Score = 87.4 bits (215), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 53/93 (56%), Positives = 62/93 (66%), Gaps = 5/93 (5%)
Query 77 DKKRKQQQQQKQQQQPNKKAKNQGGEANTSS-SGDEASYRNALVDFLKKNGPTPLASLGQ 135
DKKRK + Q NKKAK + + GD A+Y++ALVDFLKKNG TPLA+LGQ
Sbjct 222 DKKRKAVEVA---QASNKKAKPSAAPSKGAPQKGDAAAYKSALVDFLKKNGKTPLATLGQ 278
Query 136 KVKKPASLP-KMAAFLKANSDKFAVEGGKCSLK 167
KVKKPA + KMA FLKAN+ F V G CSLK
Sbjct 279 KVKKPAGVSEKMAQFLKANASTFDVSNGVCSLK 311
> pfa:PF14_0649 conserved Plasmodium protein, unknown function
Length=2558
Score = 42.0 bits (97), Expect = 0.001, Method: Composition-based stats.
Identities = 16/84 (19%), Positives = 52/84 (61%), Gaps = 0/84 (0%)
Query 24 QAAAANDDEDDEDDEDDEDDEDDDEEGSEDESGEEDEEESEEEEEAPKQQQQQDKKRKQQ 83
Q ++++D+ DE+ ++++D++++ ++DE+ +E+++ES+ E + Q +++++ +
Sbjct 2037 QNENQDENQDENQDENQDENQDENQDENQDENRDENQDESQNENQNENQDDNENEEKPNE 2096
Query 84 QQQKQQQQPNKKAKNQGGEANTSS 107
+ +++PN + EA+ +
Sbjct 2097 GSNENEEKPNDGSIENNEEADKEA 2120
Score = 40.0 bits (92), Expect = 0.003, Method: Composition-based stats.
Identities = 17/82 (20%), Positives = 46/82 (56%), Gaps = 0/82 (0%)
Query 39 DDEDDEDDDEEGSEDESGEEDEEESEEEEEAPKQQQQQDKKRKQQQQQKQQQQPNKKAKN 98
+ +++D++++ ++DE+ +E+++E+++E + + + QD+ + + Q + Q N++ N
Sbjct 2036 NQNENQDENQDENQDENQDENQDENQDENQDENRDENQDESQNENQNENQDDNENEEKPN 2095
Query 99 QGGEANTSSSGDEASYRNALVD 120
+G N D + N D
Sbjct 2096 EGSNENEEKPNDGSIENNEEAD 2117
Score = 31.2 bits (69), Expect = 1.9, Method: Composition-based stats.
Identities = 14/58 (24%), Positives = 33/58 (56%), Gaps = 0/58 (0%)
Query 29 NDDEDDEDDEDDEDDEDDDEEGSEDESGEEDEEESEEEEEAPKQQQQQDKKRKQQQQQ 86
N DE+ ++++D+ +E+ +E ++E+ E+ E S E EE P ++ + ++ +
Sbjct 2064 NQDENRDENQDESQNENQNENQDDNENEEKPNEGSNENEEKPNDGSIENNEEADKEAK 2121
> pfa:PF14_0480 conserved Plasmodium protein, unknown function
Length=1653
Score = 35.0 bits (79), Expect = 0.12, Method: Composition-based stats.
Identities = 29/96 (30%), Positives = 51/96 (53%), Gaps = 8/96 (8%)
Query 1 EEEDTKKKGKVSSKKGAAALKALQAAAANDDEDDEDDEDDEDDEDDDEEGS-EDESGEED 59
E+EDT K ++K K + +D +D+ EDD ++D E S ED+S E+D
Sbjct 986 EKEDTWK-----NQKHKGINKHKDDSHEDDSHEDDSHEDDSHEDDSHENDSHEDDSHEDD 1040
Query 60 EEESEEEEEAPKQQQQQDKKRKQQQQQKQQQQPNKK 95
E++ EE K+Q +K+ ++ ++K + NK+
Sbjct 1041 SHENDSHEE--KEQYIYNKELYEKIKKKPGKNMNKE 1074
> mmu:353326 Rtl1, 6430411K18Rik, Mar, Mart1, Mor1, Peg11; retrotransposon-like
1
Length=1744
Score = 32.0 bits (71), Expect = 0.93, Method: Composition-based stats.
Identities = 17/68 (25%), Positives = 34/68 (50%), Gaps = 5/68 (7%)
Query 28 ANDDEDDEDDEDDEDDEDDDEEGSEDESGEEDE-----EESEEEEEAPKQQQQQDKKRKQ 82
A+D+ D+ D DD + E ++G+ D+ E E P + Q+K R+Q
Sbjct 825 ADDETSDQPSSDGSDDLSESEPSELQQAGDSDQSGVFYESGARETLEPVSARMQEKARQQ 884
Query 83 QQQQKQQQ 90
++ ++Q++
Sbjct 885 EKAREQEE 892
> dre:558299 slc24a1, si:ch211-117i10.3; solute carrier family
24 (sodium/potassium/calcium exchanger), member 1; K13749 solute
carrier family 24 (sodium/potassium/calcium exchanger),
member 1
Length=724
Score = 30.4 bits (67), Expect = 2.9, Method: Composition-based stats.
Identities = 14/56 (25%), Positives = 31/56 (55%), Gaps = 2/56 (3%)
Query 48 EEGSEDESGEEDE--EESEEEEEAPKQQQQQDKKRKQQQQQKQQQQPNKKAKNQGG 101
E +++SGE D+ E++++ ++AP + +D +K + Q+ ++N GG
Sbjct 422 ENKKKEKSGEGDDQSEQTKDSKDAPPGAKDEDTNQKSEAQKDGDIAAGGGSENPGG 477
Lambda K H
0.293 0.117 0.298
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4092719652
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40