bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0160_orf4
Length=246
Score E
Sequences producing significant alignments: (Bits) Value
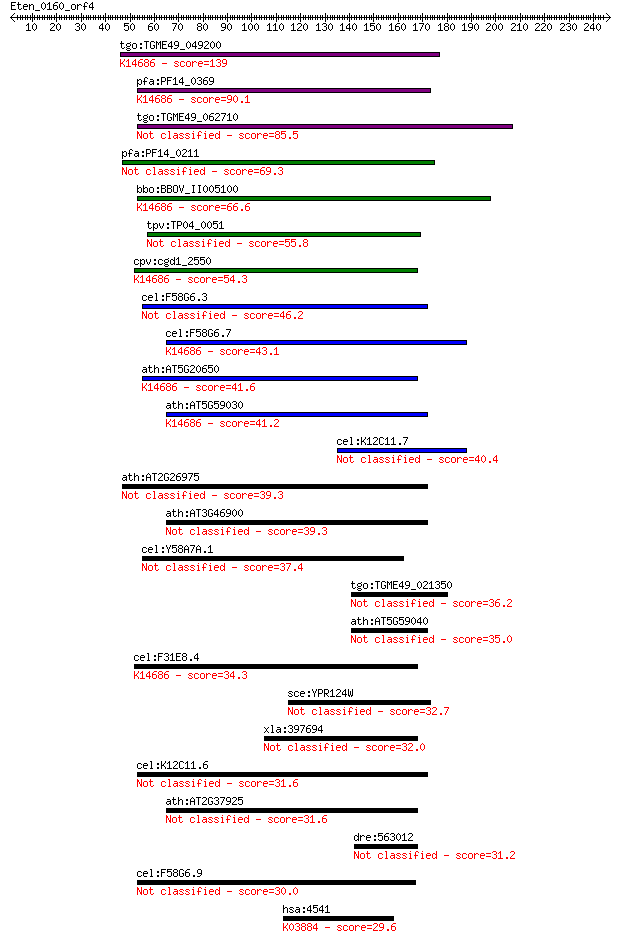
tgo:TGME49_049200 ctr copper transporter domain-containing pro... 139 1e-32
pfa:PF14_0369 copper transporter putative; K14686 solute carri... 90.1 7e-18
tgo:TGME49_062710 ctr copper transporter domain-containing pro... 85.5 2e-16
pfa:PF14_0211 Ctr copper transporter domain containing protein... 69.3 1e-11
bbo:BBOV_II005100 18.m09998; hypothetical protein; K14686 solu... 66.6 7e-11
tpv:TP04_0051 polymorphic immunodominant molecule 55.8 1e-07
cpv:cgd1_2550 copper transporter, 3 transmembrane domain, cons... 54.3 4e-07
cel:F58G6.3 hypothetical protein 46.2 1e-04
cel:F58G6.7 hypothetical protein; K14686 solute carrier family... 43.1 9e-04
ath:AT5G20650 COPT5; COPT5; copper ion transmembrane transport... 41.6 0.002
ath:AT5G59030 COPT1; COPT1 (copper transporter 1); copper ion ... 41.2 0.004
cel:K12C11.7 hypothetical protein 40.4 0.005
ath:AT2G26975 copper transporter, putative 39.3 0.012
ath:AT3G46900 COPT2; COPT2; copper ion transmembrane transport... 39.3 0.014
cel:Y58A7A.1 hypothetical protein 37.4 0.045
tgo:TGME49_021350 hypothetical protein 36.2 0.12
ath:AT5G59040 COPT3; COPT3; copper ion transmembrane transport... 35.0 0.23
cel:F31E8.4 hypothetical protein; K14686 solute carrier family... 34.3 0.48
sce:YPR124W CTR1; Ctr1p 32.7 1.3
xla:397694 slc31a2, copt2, ctr2, xem1; solute carrier family 3... 32.0 2.2
cel:K12C11.6 hypothetical protein 31.6 3.0
ath:AT2G37925 COPT4; COPT4; copper ion transmembrane transport... 31.6 3.1
dre:563012 sb:cb797; si:ch211-282j22.4; K14687 solute carrier ... 31.2 3.6
cel:F58G6.9 hypothetical protein 30.0 7.9
hsa:4541 ND6, MTND6; NADH dehydrogenase, subunit 6 (complex I)... 29.6 9.7
> tgo:TGME49_049200 ctr copper transporter domain-containing protein
; K14686 solute carrier family 31 (copper transporter),
member 1
Length=235
Score = 139 bits (350), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 67/131 (51%), Positives = 88/131 (67%), Gaps = 0/131 (0%)
Query 46 LNKDGFPLPMSFEASPEVLLLFSWWHARTNAQYAVTCVCCIIFGFISIALKGLRRISDVR 105
L+ DG PLPM+FE S V+ LF W T Q+A CV I GFI + LK +RR +
Sbjct 67 LSIDGMPLPMAFEVSTRVIYLFEDWPTETTTQFAGACVATCILGFICVILKVVRRYVEKS 126
Query 106 LAMMENRSKPTLLFGCFPVFHNAIRGCVTFLNYSWDYMLMLVAMTFNVGIFVSMLGGMAL 165
L EN K L+FG FP++ N++R V F+NYSWDYMLML++MTFNVGIF+S+L G+AL
Sbjct 127 LVSQENVGKTKLIFGSFPLYSNSVRFLVAFVNYSWDYMLMLLSMTFNVGIFLSLLLGIAL 186
Query 166 GFLTIGRFLDL 176
GFL +G + +
Sbjct 187 GFLFLGDLMSV 197
> pfa:PF14_0369 copper transporter putative; K14686 solute carrier
family 31 (copper transporter), member 1
Length=235
Score = 90.1 bits (222), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 44/121 (36%), Positives = 76/121 (62%), Gaps = 5/121 (4%)
Query 53 LPMSFEASPEVLLLFSWWHARTNAQYAVTCVCCIIFGFISIALKGLRRISDVRLAMMENR 112
+PMSF+ + ++LF+ W ++ Y ++ V C FG IS+ K +R +V A+ +
Sbjct 91 MPMSFQLTTHTIILFNKWETKSALSYYISLVLCFFFGIISVGFKVVRL--NVEQAL--PK 146
Query 113 SKPTLLFGCFPVF-HNAIRGCVTFLNYSWDYMLMLVAMTFNVGIFVSMLGGMALGFLTIG 171
++ T +F +F +N+ R ++F+ YSWDY+LML+ MTFNVG+FV+++ G++ GF G
Sbjct 147 TEDTNIFKSLVLFKNNSYRMLLSFVIYSWDYLLMLIVMTFNVGLFVAVVLGLSFGFFIFG 206
Query 172 R 172
Sbjct 207 N 207
> tgo:TGME49_062710 ctr copper transporter domain-containing protein
Length=375
Score = 85.5 bits (210), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 52/178 (29%), Positives = 83/178 (46%), Gaps = 33/178 (18%)
Query 53 LPMSFEASPEVLLLFSWWHARTNAQYAVTCVCCIIFGFISIALKGLRRISDVRLAMMENR 112
+PMSF+ S ++LF W QY ++ + C++ G +S+ LK LR + LA +
Sbjct 206 MPMSFQNSLHTVILFHSWETLERWQYVLSLLTCVVLGMLSVVLKVLRLRLEFFLAKRDRA 265
Query 113 SKPTL------------------------LFGCFPVFHNAIRGCVTFLNYSWDYMLMLVA 148
++ L G FP+ N+ R F+ Y +DY+LML+
Sbjct 266 AEDAQRVEKLKEKEGQSSAASPSSAIVERLCGNFPLKQNSWRMLEAFVIYGYDYLLMLIV 325
Query 149 MTFNVGIFVSMLGGMALGFLTIGRFLDLPFEPAKVSGGCECNETLSCGCHKGRPCTCA 206
MT+NVG+F ++ GG+ALGF G L + E + S + ++G PC C
Sbjct 326 MTYNVGLFFAVTGGLALGFFCFGHLLRIQAEKEENSLEED---------YRGDPCCCG 374
> pfa:PF14_0211 Ctr copper transporter domain containing protein,
putative
Length=160
Score = 69.3 bits (168), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 39/128 (30%), Positives = 68/128 (53%), Gaps = 2/128 (1%)
Query 47 NKDGFPLPMSFEASPEVLLLFSWWHARTNAQYAVTCVCCIIFGFISIALKGLRRISDVRL 106
N DG LPM F + + +LF + + Q+ + CII GF S+ +K L++ +
Sbjct 35 NDDGVMLPMYFSNNENIKMLFDIFQVKNRYQFIFCNILCIIMGFFSVYIKVLKKRLHHNV 94
Query 107 AMMENRSKPTLLFGCFPVFHNAIRGCVTFLNYSWDYMLMLVAMTFNVGIFVSMLGGMALG 166
+ + + + P N G ++FL+Y+ DY+LML+ MTFN IF+S++ G++
Sbjct 95 QKVADGGDGSYV-NMSPC-QNVNYGFLSFLHYTIDYLLMLIVMTFNPYIFLSIMTGLSSA 152
Query 167 FLTIGRFL 174
+L G +
Sbjct 153 YLFYGHLI 160
> bbo:BBOV_II005100 18.m09998; hypothetical protein; K14686 solute
carrier family 31 (copper transporter), member 1
Length=297
Score = 66.6 bits (161), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 42/145 (28%), Positives = 69/145 (47%), Gaps = 12/145 (8%)
Query 53 LPMSFEASPEVLLLFSWWHARTNAQYAVTCVCCIIFGFISIALKGLRRISDVRLAMMENR 112
+PM FE + + ++LF +W T QYAV+ + +++ LK R + L N
Sbjct 163 MPMYFENTVKTVILFHFWKTTTGTQYAVSLFFIFVLSLMTVFLKAFRNKLNCALLQRPNG 222
Query 113 SKPTLLFGCFPVFHNAIRGCVTFLNYSWDYMLMLVAMTFNVGIFVSMLGGMALGFLTIGR 172
PT+ +G + + VTF+ D+ +MLV MTFNVGI + + ALG++
Sbjct 223 YHPTVKYGIMYI----LAFVVTFM----DFAMMLVVMTFNVGIVLVVCSAYALGYI---- 270
Query 173 FLDLPFEPAKVSGGCECNETLSCGC 197
F P ++ + G C + C
Sbjct 271 FTCCPLGLSEAANGSRCTQECPADC 295
> tpv:TP04_0051 polymorphic immunodominant molecule
Length=480
Score = 55.8 bits (133), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 31/112 (27%), Positives = 58/112 (51%), Gaps = 9/112 (8%)
Query 57 FEASPEVLLLFSWWHARTNAQYAVTCVCCIIFGFISIALKGLRRISDVRLAMMENRSKPT 116
F + +V ++F WW QYA+T + F +S LK R + + ++ +
Sbjct 351 FTNTHKVTVIFHWWLCEKPWQYALTLLTLFGFALLSPCLKAYREV-------LRAKAVRS 403
Query 117 LLFGCFPVFHNAIRGCVTFLNYSWDYMLMLVAMTFNVGIFVSMLGGMALGFL 168
+F C + + + Y+ D++LMLV MTFNVG+F +++ G ++G++
Sbjct 404 FIFDC--LLTHLFLFLIALCAYALDFLLMLVVMTFNVGVFFAVILGYSVGYV 453
> cpv:cgd1_2550 copper transporter, 3 transmembrane domain, conserved
in metazoa and apiacomplexa ; K14686 solute carrier
family 31 (copper transporter), member 1
Length=178
Score = 54.3 bits (129), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 32/118 (27%), Positives = 63/118 (53%), Gaps = 10/118 (8%)
Query 52 PLPMSFEASPEVLLLFSWWHARTNAQYAVTCVCCIIFGFISIALKGLRR--ISDVRLAMM 109
+ M+F S E ++LF W Y ++C+ I+ G ++ + + + I +++ +
Sbjct 44 AMQMTFHQSFESVILFESWRTSNRFDYFISCLFIILMGCFTMFISSINKKYIKEIKKNRV 103
Query 110 ENRSKPTLLFGCFPVFHNAIRGCVTFLNYSWDYMLMLVAMTFNVGIFVSMLGGMALGF 167
E+ + G + N + +T L Y Y+LML+AMTFN G+F S++ G+++G+
Sbjct 104 EHEN-----LGIKVICTNVL---LTILYYFMHYLLMLIAMTFNWGLFFSVIIGLSIGY 153
> cel:F58G6.3 hypothetical protein
Length=134
Score = 46.2 bits (108), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 32/117 (27%), Positives = 57/117 (48%), Gaps = 4/117 (3%)
Query 55 MSFEASPEVLLLFSWWHARTNAQYAVTCVCCIIFGFISIALKGLRRISDVRLAMMENRSK 114
M F P+ +LFS W+ + + C+ I G I A+K RR+ R + + S
Sbjct 18 MWFHTKPQDTVLFSTWNITSAGKMVWACILVAIAGIILEAIKYNRRLIQKRQSPSKKESY 77
Query 115 PTLLFGCFPVFHNAIRGCVTFLNYSWDYMLMLVAMTFNVGIFVSMLGGMALGFLTIG 171
+ L F + F+ + Y LML+ MTF++ + ++++ G+++GFL G
Sbjct 78 ISRLLSTMHFFQT----FLFFVQLGFSYCLMLIFMTFSIWLGLAVVIGLSIGFLIFG 130
> cel:F58G6.7 hypothetical protein; K14686 solute carrier family
31 (copper transporter), member 1
Length=166
Score = 43.1 bits (100), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 36/150 (24%), Positives = 62/150 (41%), Gaps = 30/150 (20%)
Query 65 LLFSWWHARTNAQYAVTCVCCIIFGFISIALKGLRRISDVRLAMMENR------------ 112
+LF W Y +C+ + F ALK R ++ ++E +
Sbjct 18 ILFREWKPLNTTAYVFSCIEIFLIAFCLEALKFGRTKLSPKVKIVEKKVDCCCSTEKDGL 77
Query 113 ---------SKPTLLFGCFP------VFHNAIRGCVTFLNYSWDYMLMLVAMTFNVGIFV 157
++ T+ F FH A + F+ + DY LMLV+MT+N IF+
Sbjct 78 WNIPETIPLTQKTVTLAPFTRDSLISKFHMA-SSLLVFVQHFIDYSLMLVSMTYNWPIFL 136
Query 158 SMLGGMALGFLTIGRFLDLPFEPAKVSGGC 187
S+L G G+ +G + + E ++ +G C
Sbjct 137 SLLAGHTTGYFFLGPMMTV--EESEAAGSC 164
> ath:AT5G20650 COPT5; COPT5; copper ion transmembrane transporter/
high affinity copper ion transmembrane transporter; K14686
solute carrier family 31 (copper transporter), member 1
Length=146
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 32/122 (26%), Positives = 55/122 (45%), Gaps = 9/122 (7%)
Query 55 MSFEASPEVLLLFSWWHARTNAQYAVTCVCCIIFGFISIALKGLR----RISDVRLAMME 110
M+F + +LF +W + Y +T + C +F L+ R +S R A
Sbjct 4 MTFYWGIKATILFDFWKTDSWLSYILTLIACFVFSAFYQYLENRRIQFKSLSSSRRAPPP 63
Query 111 NRSKPTLLFGCFPV--FHNAIRGCVTFL---NYSWDYMLMLVAMTFNVGIFVSMLGGMAL 165
RS + P +A + L N + Y+LML AM+FN G+F++++ G+
Sbjct 64 PRSSSGVSAPLIPKSGTRSAAKAASVLLFGVNAAIGYLLMLAAMSFNGGVFIAIVVGLTA 123
Query 166 GF 167
G+
Sbjct 124 GY 125
> ath:AT5G59030 COPT1; COPT1 (copper transporter 1); copper ion
transmembrane transporter; K14686 solute carrier family 31
(copper transporter), member 1
Length=170
Score = 41.2 bits (95), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 30/107 (28%), Positives = 52/107 (48%), Gaps = 13/107 (12%)
Query 65 LLFSWWHARTNAQYAVTCVCCIIFGFISIALKGLRRISDVRLAMMENRSKPTLLFGCFPV 124
+LFS W ++ YA+ C I F+++ + L S +R + ++ ++ L
Sbjct 54 VLFSGWPGTSSGMYAL---CLIFVFFLAVLTEWLAHSSLLRGSTGDSANRAAGL------ 104
Query 125 FHNAIRGCVTFLNYSWDYMLMLVAMTFNVGIFVSMLGGMALGFLTIG 171
I+ V L Y++ML M+FN G+F+ L G A+GF+ G
Sbjct 105 ----IQTAVYTLRIGLAYLVMLAVMSFNAGVFLVALAGHAVGFMLFG 147
> cel:K12C11.7 hypothetical protein
Length=166
Score = 40.4 bits (93), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 21/53 (39%), Positives = 32/53 (60%), Gaps = 2/53 (3%)
Query 135 FLNYSWDYMLMLVAMTFNVGIFVSMLGGMALGFLTIGRFLDLPFEPAKVSGGC 187
FL DY LMLVAMT+N +F S+L G A+G+ +G + + + + +G C
Sbjct 114 FLQNFVDYSLMLVAMTYNYPLFFSLLAGHAIGYFFVGPLMTV--KEVENTGNC 164
> ath:AT2G26975 copper transporter, putative
Length=145
Score = 39.3 bits (90), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 30/125 (24%), Positives = 55/125 (44%), Gaps = 15/125 (12%)
Query 47 NKDGFPLPMSFEASPEVLLLFSWWHARTNAQYAVTCVCCIIFGFISIALKGLRRISDVRL 106
N + + M+F +LFS W + Y +C I+ +++ ++ L S +R
Sbjct 19 NSNMIMMHMTFFWGKNTEILFSGWPGTSLGMY---VLCLIVVFLLAVIVEWLAHSSILRG 75
Query 107 AMMENRSKPTLLFGCFPVFHNAIRGCVTFLNYSWDYMLMLVAMTFNVGIFVSMLGGMALG 166
+R+K ++ V L Y++ML M+FN G+F+ + G A+G
Sbjct 76 RGSTSRAK------------GLVQTAVYTLKTGLAYLVMLAVMSFNGGVFIVAIAGFAVG 123
Query 167 FLTIG 171
F+ G
Sbjct 124 FMLFG 128
> ath:AT3G46900 COPT2; COPT2; copper ion transmembrane transporter/
high affinity copper ion transmembrane transporter
Length=158
Score = 39.3 bits (90), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 26/107 (24%), Positives = 47/107 (43%), Gaps = 15/107 (14%)
Query 65 LLFSWWHARTNAQYAVTCVCCIIFGFISIALKGLRRISDVRLAMMENRSKPTLLFGCFPV 124
+LFS W ++ YA+ C I+ +++ + L +R++ NR+
Sbjct 42 VLFSGWPGTSSGMYAL---CLIVIFLLAVIAEWLAHSPILRVSGSTNRAA---------- 88
Query 125 FHNAIRGCVTFLNYSWDYMLMLVAMTFNVGIFVSMLGGMALGFLTIG 171
+ V L Y++ML M+FN G+F+ + G +GF G
Sbjct 89 --GLAQTAVYTLKTGLSYLVMLAVMSFNAGVFIVAIAGYGVGFFLFG 133
> cel:Y58A7A.1 hypothetical protein
Length=130
Score = 37.4 bits (85), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 27/108 (25%), Positives = 44/108 (40%), Gaps = 2/108 (1%)
Query 55 MSFEASPEVLLLFSWWHARTNAQYAVTCVCCIIFGFISIALKGLRRISDVRLAMMENRSK 114
MSF E +LF +W T AV C ++ F+ L+ R + + +
Sbjct 1 MSFHFGTEETILFDFWKTETAVGIAVACFITVLLAFLMETLRFFRDYRKAQTQLHQPPIS 60
Query 115 PTLLFGCFPVFHNAIRGCVTFLNYSWDYMLMLVAMTFNVGI-FVSMLG 161
P P + I + + Y LML+ MTFN + F +++G
Sbjct 61 PEDRLKRSPQL-DLIDPLLQLFQLTIAYFLMLIFMTFNAYLCFFTVVG 107
> tgo:TGME49_021350 hypothetical protein
Length=300
Score = 36.2 bits (82), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 17/43 (39%), Positives = 27/43 (62%), Gaps = 4/43 (9%)
Query 141 DYMLMLVAMTFNVGIFVSMLGGMALGFLTIG----RFLDLPFE 179
D+ LMLV MTFN G+F++++ G+A G I + LP++
Sbjct 249 DWSLMLVCMTFNAGLFLTVVAGVAAGRTLINSKTRKLWALPWQ 291
> ath:AT5G59040 COPT3; COPT3; copper ion transmembrane transporter/
high affinity copper ion transmembrane transporter
Length=151
Score = 35.0 bits (79), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 21/31 (67%), Gaps = 0/31 (0%)
Query 141 DYMLMLVAMTFNVGIFVSMLGGMALGFLTIG 171
Y++ML M+FN G+FV+ + G LGF+ G
Sbjct 102 SYLVMLAVMSFNGGVFVAAMAGFGLGFMIFG 132
> cel:F31E8.4 hypothetical protein; K14686 solute carrier family
31 (copper transporter), member 1
Length=162
Score = 34.3 bits (77), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 31/148 (20%), Positives = 62/148 (41%), Gaps = 36/148 (24%)
Query 52 PLPMSFEASPEVLLLFSWWHARTNAQYAVTCVCCIIFGFISIALKGLR------------ 99
+ M+ +LFSWW + + AV+ + + + A+K R
Sbjct 2 DMDMTLHFGEREKILFSWWKTGSLSGMAVSMLITFLLCILYEAIKSFRYFLAVWNNQKRQ 61
Query 100 ------RISDVRLAMMENRSKPTL--------------LFGCFPVFHNAIRGCVTFLNYS 139
I++ + + +N S+ ++ LF + + A+ G L Y+
Sbjct 62 QRHAEASITNPQNSGGDNISEDSIHIAPLVQLSGFTKRLFTSYRLAQGALYGLQALLAYT 121
Query 140 WDYMLMLVAMTFNVGIFVSMLGGMALGF 167
LML+AMT+N+ + +S++ G A+G+
Sbjct 122 ----LMLIAMTYNMNLILSIVVGEAVGY 145
> sce:YPR124W CTR1; Ctr1p
Length=406
Score = 32.7 bits (73), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 31/63 (49%), Gaps = 5/63 (7%)
Query 115 PTLLFGCF-----PVFHNAIRGCVTFLNYSWDYMLMLVAMTFNVGIFVSMLGGMALGFLT 169
P LL F +FH+ IR + F + YMLML M+F + +++ G+AL +
Sbjct 221 PNLLSDIFVPSLMDLFHDIIRAFLVFTSTMIIYMLMLATMSFVLTYVFAVITGLALSEVF 280
Query 170 IGR 172
R
Sbjct 281 FNR 283
> xla:397694 slc31a2, copt2, ctr2, xem1; solute carrier family
31 (copper transporters), member 2; K14687 solute carrier family
31 (copper transporter), member 2
Length=172
Score = 32.0 bits (71), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 18/63 (28%), Positives = 33/63 (52%), Gaps = 4/63 (6%)
Query 105 RLAMMENRSKPTLLFGCFPVFHNAIRGCVTFLNYSWDYMLMLVAMTFNVGIFVSMLGGMA 164
RL++ E +P+ + F + +R L Y +LML M++N IF++++ G
Sbjct 98 RLSVTEEHIQPSSRWWFLHSFLSLLRMVQVVLGY----LLMLCVMSYNAAIFIAVILGSG 153
Query 165 LGF 167
LG+
Sbjct 154 LGY 156
> cel:K12C11.6 hypothetical protein
Length=132
Score = 31.6 bits (70), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 32/123 (26%), Positives = 58/123 (47%), Gaps = 9/123 (7%)
Query 53 LPMSFEASPEVLLLFSWWHARTNAQYAVTCVCCIIFGFISIALKGLR-RISDVRLAMME- 110
+ M F + +LF W+ C ++ G + +K LR +I E
Sbjct 9 MHMWFHTKTQDTVLFKTWNVTDTPTMVWVCCIIVVAGILLELIKFLRWKIEKWHKNRDEL 68
Query 111 -NRSKPTLLFGCFPVFHNAIRGCVTFL-NYSWDYMLMLVAMTFNVGIFVSMLGGMALGFL 168
+RS + LF P+ I + F+ S+ Y+LML+ MTF+V + ++++ G+ +G+L
Sbjct 69 VSRSYISRLFS--PI---HIGQTILFMVQLSFSYILMLLFMTFSVWLGIAVVVGLGIGYL 123
Query 169 TIG 171
G
Sbjct 124 AFG 126
> ath:AT2G37925 COPT4; COPT4; copper ion transmembrane transporter/
high affinity copper ion transmembrane transporter
Length=145
Score = 31.6 bits (70), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 25/103 (24%), Positives = 46/103 (44%), Gaps = 13/103 (12%)
Query 65 LLFSWWHARTNAQYAVTCVCCIIFGFISIALKGLRRISDVRLAMMENRSKPTLLFGCFPV 124
+LFS W YA+ + F++ + L R SD ++ G +
Sbjct 42 VLFSGWPGSDRGMYALALIFVFFLAFLA---EWLARCSDA----------SSIKQGADKL 88
Query 125 FHNAIRGCVTFLNYSWDYMLMLVAMTFNVGIFVSMLGGMALGF 167
A R + + + Y+++L ++FN G+F++ + G ALGF
Sbjct 89 AKVAFRTAMYTVKSGFSYLVILAVVSFNGGVFLAAIFGHALGF 131
> dre:563012 sb:cb797; si:ch211-282j22.4; K14687 solute carrier
family 31 (copper transporter), member 2
Length=171
Score = 31.2 bits (69), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 12/26 (46%), Positives = 19/26 (73%), Gaps = 0/26 (0%)
Query 142 YMLMLVAMTFNVGIFVSMLGGMALGF 167
YMLML M++N+ IF+ ++ G LG+
Sbjct 136 YMLMLCVMSYNIWIFLGVITGSVLGY 161
> cel:F58G6.9 hypothetical protein
Length=156
Score = 30.0 bits (66), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 29/124 (23%), Positives = 50/124 (40%), Gaps = 10/124 (8%)
Query 53 LPMSFEASPEVLLLFSWWHARTNAQYAVTCVCCIIFGFISIALKGLRRISDVRLAMMENR 112
+ M + E +LF W TC G + ALK R ++ R+ + +
Sbjct 21 MWMWYHVDVEDTVLFKSWTVFDAGTMVWTCFVVAAAGILLEALKYARWATEERMKIDQEN 80
Query 113 SKPTLLFGCFPV------FHNAIRGCVTFLNYSWD----YMLMLVAMTFNVGIFVSMLGG 162
+G + ++ R + L + W Y+LM V M F+V I +S+ G
Sbjct 81 VDSKTKYGGIKIPGKSEKYNFWKRHIIDSLYHFWQLLLAYILMNVYMVFSVYICLSLCFG 140
Query 163 MALG 166
+A+G
Sbjct 141 LAIG 144
> hsa:4541 ND6, MTND6; NADH dehydrogenase, subunit 6 (complex
I); K03884 NADH dehydrogenase I subunit 6 [EC:1.6.5.3]
Length=174
Score = 29.6 bits (65), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 26/45 (57%), Gaps = 0/45 (0%)
Query 113 SKPTLLFGCFPVFHNAIRGCVTFLNYSWDYMLMLVAMTFNVGIFV 157
SKP+ ++G + + + GCV LN+ YM ++V + + G+ V
Sbjct 21 SKPSPIYGGLVLIVSGVVGCVIILNFGGGYMGLMVFLIYLGGMMV 65
Lambda K H
0.328 0.140 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8700848608
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40