bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0220_orf3
Length=236
Score E
Sequences producing significant alignments: (Bits) Value
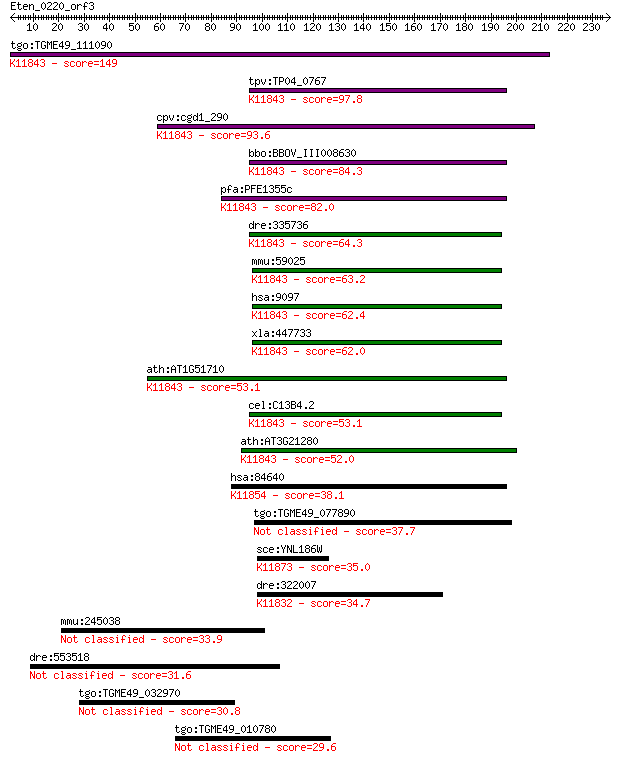
tgo:TGME49_111090 ubiquitin carboxyl-terminal hydrolase, putat... 149 1e-35
tpv:TP04_0767 ubiquitin carboxyl-terminal hydrolase (EC:3.1.2.... 97.8 3e-20
cpv:cgd1_290 Ub6p like ubiquitin at N-terminus and ubiquitin C... 93.6 6e-19
bbo:BBOV_III008630 17.m07754; ubiquitin carboxyl-terminal hydr... 84.3 3e-16
pfa:PFE1355c ubiquitin carboxyl-terminal hydrolase, putative (... 82.0 2e-15
dre:335736 usp14, wu:fk63d09, zgc:55949; ubiquitin specific pr... 64.3 4e-10
mmu:59025 Usp14, 2610005K12Rik, 2610037B11Rik, AW107924, C7876... 63.2 7e-10
hsa:9097 USP14, TGT; ubiquitin specific peptidase 14 (tRNA-gua... 62.4 1e-09
xla:447733 usp14, MGC81945; ubiquitin specific peptidase 14 (t... 62.0 2e-09
ath:AT1G51710 UBP6; UBP6 (UBIQUITIN-SPECIFIC PROTEASE 6); calm... 53.1 9e-07
cel:C13B4.2 usp-14; Ubiquitin Specific Protease family member ... 53.1 9e-07
ath:AT3G21280 UBP7; UBP7 (UBIQUITIN-SPECIFIC PROTEASE 7); ubiq... 52.0 2e-06
hsa:84640 USP38, FLJ35970, HP43.8KD, KIAA1891; ubiquitin speci... 38.1 0.031
tgo:TGME49_077890 hypothetical protein 37.7 0.034
sce:YNL186W UBP10, DOT4; Ubiquitin-specific protease that deub... 35.0 0.21
dre:322007 usp1, wu:fb43a02, zgc:55962; ubiquitin specific pro... 34.7 0.29
mmu:245038 Dclk3, BC056929, C730036H08, Dcamkl3; doublecortin-... 33.9 0.46
dre:553518 Phf3; si:dkey-188i13.1 31.6 2.7
tgo:TGME49_032970 hypothetical protein 30.8 3.9
tgo:TGME49_010780 ubiquitin carboxyl-terminal hydrolase, putat... 29.6 8.8
> tgo:TGME49_111090 ubiquitin carboxyl-terminal hydrolase, putative
(EC:3.1.2.15 3.1.3.16); K11843 ubiquitin carboxyl-terminal
hydrolase 14 [EC:3.1.2.15]
Length=607
Score = 149 bits (375), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 93/216 (43%), Positives = 118/216 (54%), Gaps = 38/216 (17%)
Query 1 FKVGRAIVALRREREASSGGAEKTTNGTASTESGAPAADGAAAATAAPEAAAVPAATNGS 60
++GRA+VALRREREA G + + T T S AA T E + V ++ GS
Sbjct 404 LRIGRAVVALRREREA---GGHSSDSKTEETNSKTEPTCADAAMTEKNEDSNV--SSTGS 458
Query 61 GAAHSASAPAAEAAEAEAKKKETELLRLAGKPCPTGTYQLLSVVTHQGRYADSGHYVGWA 120
T + R TG +QLL +VTHQGR+ADSGHYVGW
Sbjct 459 ----------------------TRVYR-------TGFFQLLGIVTHQGRHADSGHYVGWT 489
Query 121 RAG--DAGTLQGPPTAQSEDKEEEEPAAPSGPAAKRRKN--VDMWRKFDDDKVSEVPWDS 176
+ + ++ + ++E + PSG A K++K VDMW KFDDDKVSE PWD
Sbjct 490 KKDQREPRVIEKEKKEKEREEELQREENPSGMAVKKKKASPVDMWVKFDDDKVSETPWDQ 549
Query 177 IDLAGGRSDYHVAYLLLMKHILVVPTEEELLAVAKQ 212
IDLAGGRSDYH+AYLLL + ILV TEEE+ V K
Sbjct 550 IDLAGGRSDYHIAYLLLFREILVDATEEEVKTVEKN 585
> tpv:TP04_0767 ubiquitin carboxyl-terminal hydrolase (EC:3.1.2.15);
K11843 ubiquitin carboxyl-terminal hydrolase 14 [EC:3.1.2.15]
Length=526
Score = 97.8 bits (242), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 46/101 (45%), Positives = 59/101 (58%), Gaps = 3/101 (2%)
Query 95 TGTYQLLSVVTHQGRYADSGHYVGWARAGDAGTLQGPPTAQSEDKEEEEPAAPSGPAAKR 154
TG Y+L+S+VTHQGR AD+GHY+ W + + P + +D + + K+
Sbjct 423 TGKYELISIVTHQGRTADAGHYICWTKDSREYPTKNPSNSNEKDMDNQRDGEKESKKKKQ 482
Query 155 RKNVDMWRKFDDDKVSEVPWDSIDLAGGRSDYHVAYLLLMK 195
D W KFDDD VSE W S DL GGRSDYH+A LLL K
Sbjct 483 E---DRWIKFDDDVVSEQDWGSFDLCGGRSDYHIAVLLLYK 520
> cpv:cgd1_290 Ub6p like ubiquitin at N-terminus and ubiquitin
C terminal hydrolase at the C-terminus ; K11843 ubiquitin carboxyl-terminal
hydrolase 14 [EC:3.1.2.15]
Length=499
Score = 93.6 bits (231), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 56/148 (37%), Positives = 71/148 (47%), Gaps = 36/148 (24%)
Query 59 GSGAAHSASAPAAEAAEAEAKKKETELLRLAGKPCPTGTYQLLSVVTHQGRYADSGHYVG 118
G + + A E + E +K ETEL CPTG Y+L VVTHQGR ADSGHYV
Sbjct 383 GRDISEKKKSKAIEKPDQEEQKMETELY----TDCPTGVYELECVVTHQGRTADSGHYVA 438
Query 119 WARAGDAGTLQGPPTAQSEDKEEEEPAAPSGPAAKRRKNVDMWRKFDDDKVSEVPWDSID 178
W + D+E KFDDDKVS + D
Sbjct 439 WRYCPN-------------DRE-------------------YIIKFDDDKVSRIKAKDAD 466
Query 179 LAGGRSDYHVAYLLLMKHILVVPTEEEL 206
L+GGRSDYH+A +LL K ++ +EEE+
Sbjct 467 LSGGRSDYHIAVMLLYKKTVIKASEEEM 494
> bbo:BBOV_III008630 17.m07754; ubiquitin carboxyl-terminal hydrolase
family protein; K11843 ubiquitin carboxyl-terminal hydrolase
14 [EC:3.1.2.15]
Length=496
Score = 84.3 bits (207), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 45/101 (44%), Positives = 54/101 (53%), Gaps = 24/101 (23%)
Query 95 TGTYQLLSVVTHQGRYADSGHYVGWARAGDAGTLQGPPTAQSEDKEEEEPAAPSGPAAKR 154
TG YQL ++VTHQGR AD GHYV WA+ P +E +
Sbjct 402 TGKYQLEAIVTHQGRSADGGHYVCWAK---------DPREDTEGSND------------- 439
Query 155 RKNVDMWRKFDDDKVSEVPWDSIDLAGGRSDYHVAYLLLMK 195
KN D W FDDDKV+E W + DL GGR D+H+A LLL K
Sbjct 440 -KN-DQWLMFDDDKVTEYRWGNFDLCGGRGDFHIAVLLLYK 478
> pfa:PFE1355c ubiquitin carboxyl-terminal hydrolase, putative
(EC:3.1.2.15); K11843 ubiquitin carboxyl-terminal hydrolase
14 [EC:3.1.2.15]
Length=605
Score = 82.0 bits (201), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 44/112 (39%), Positives = 59/112 (52%), Gaps = 16/112 (14%)
Query 84 ELLRLAGKPCPTGTYQLLSVVTHQGRYADSGHYVGWARAGDAGTLQGPPTAQSEDKEEEE 143
EL+ L PTG Y+L+SV+TH+GR +SGHY+ W + + Q+E
Sbjct 493 ELIEL-----PTGEYELISVITHKGRNEESGHYIAWKKMKKFFS-SNSNIDQNE------ 540
Query 144 PAAPSGPAAKRRKNVDMWRKFDDDKVSEVPWDSIDLAGGRSDYHVAYLLLMK 195
S + N +W K DDDKVS + SID GG SDY++A LLL K
Sbjct 541 ----SSNKKTKNANDSLWLKMDDDKVSTHKFSSIDFYGGCSDYNIAVLLLYK 588
> dre:335736 usp14, wu:fk63d09, zgc:55949; ubiquitin specific
protease 14 (tRNA-guanine transglycosylase) (EC:3.4.19.12);
K11843 ubiquitin carboxyl-terminal hydrolase 14 [EC:3.1.2.15]
Length=489
Score = 64.3 bits (155), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 39/100 (39%), Positives = 51/100 (51%), Gaps = 36/100 (36%)
Query 95 TGTYQLLSVVTHQGRYADSGHYVGWARAGDAGTLQGPPTAQSEDKEEEEPAAPSGPAAKR 154
+G Y+L +V+THQGR + SGHYV W KR
Sbjct 415 SGYYELQAVLTHQGRSSSSGHYVAWV--------------------------------KR 442
Query 155 RKNVDMWRKFDDDKVSEV-PWDSIDLAGGRSDYHVAYLLL 193
++ D W KFDDDKVS V P D + L+GG D+H+AY+LL
Sbjct 443 KE--DEWVKFDDDKVSVVTPEDILKLSGG-GDWHIAYVLL 479
> mmu:59025 Usp14, 2610005K12Rik, 2610037B11Rik, AW107924, C78769,
ax; ubiquitin specific peptidase 14 (EC:3.4.19.12); K11843
ubiquitin carboxyl-terminal hydrolase 14 [EC:3.1.2.15]
Length=458
Score = 63.2 bits (152), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 40/99 (40%), Positives = 48/99 (48%), Gaps = 36/99 (36%)
Query 96 GTYQLLSVVTHQGRYADSGHYVGWARAGDAGTLQGPPTAQSEDKEEEEPAAPSGPAAKRR 155
G Y L +V+THQGR + SGHYV W R R
Sbjct 380 GYYDLQAVLTHQGRSSSSGHYVSWVR---------------------------------R 406
Query 156 KNVDMWRKFDDDKVSEV-PWDSIDLAGGRSDYHVAYLLL 193
K D W KFDDDKVS V P D + L+GG D+H+AY+LL
Sbjct 407 KQ-DEWIKFDDDKVSIVTPEDILRLSGG-GDWHIAYVLL 443
> hsa:9097 USP14, TGT; ubiquitin specific peptidase 14 (tRNA-guanine
transglycosylase) (EC:3.4.19.12); K11843 ubiquitin carboxyl-terminal
hydrolase 14 [EC:3.1.2.15]
Length=459
Score = 62.4 bits (150), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 39/99 (39%), Positives = 49/99 (49%), Gaps = 36/99 (36%)
Query 96 GTYQLLSVVTHQGRYADSGHYVGWARAGDAGTLQGPPTAQSEDKEEEEPAAPSGPAAKRR 155
G Y L +V+THQGR + SGHYV W KR+
Sbjct 381 GYYDLQAVLTHQGRSSSSGHYVSWV--------------------------------KRK 408
Query 156 KNVDMWRKFDDDKVSEV-PWDSIDLAGGRSDYHVAYLLL 193
+ D W KFDDDKVS V P D + L+GG D+H+AY+LL
Sbjct 409 Q--DEWIKFDDDKVSIVTPEDILRLSGG-GDWHIAYVLL 444
> xla:447733 usp14, MGC81945; ubiquitin specific peptidase 14
(tRNA-guanine transglycosylase) (EC:3.1.2.15); K11843 ubiquitin
carboxyl-terminal hydrolase 14 [EC:3.1.2.15]
Length=489
Score = 62.0 bits (149), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 38/99 (38%), Positives = 50/99 (50%), Gaps = 36/99 (36%)
Query 96 GTYQLLSVVTHQGRYADSGHYVGWARAGDAGTLQGPPTAQSEDKEEEEPAAPSGPAAKRR 155
G Y+L +V+THQGR + SGHY+ W + R
Sbjct 411 GYYELQAVLTHQGRSSSSGHYLSWVK---------------------------------R 437
Query 156 KNVDMWRKFDDDKVSEV-PWDSIDLAGGRSDYHVAYLLL 193
K+ D W KFDDDKVS V P D + L+GG D+H+AY+LL
Sbjct 438 KH-DEWIKFDDDKVSIVSPEDILRLSGG-GDWHIAYVLL 474
> ath:AT1G51710 UBP6; UBP6 (UBIQUITIN-SPECIFIC PROTEASE 6); calmodulin
binding / ubiquitin-specific protease; K11843 ubiquitin
carboxyl-terminal hydrolase 14 [EC:3.1.2.15]
Length=443
Score = 53.1 bits (126), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 40/141 (28%), Positives = 59/141 (41%), Gaps = 48/141 (34%)
Query 55 AATNGSGAAHSASAPAAEAAEAEAKKKETELLRLAGKPCPTGTYQLLSVVTHQGRYADSG 114
A+ NGSG + + + ++E KET + TG Y L++V+TH+GR ADSG
Sbjct 345 ASANGSGESSTVNPQEGTSSE-----KETHM---------TGIYDLVAVLTHKGRSADSG 390
Query 115 HYVGWARAGDAGTLQGPPTAQSEDKEEEEPAAPSGPAAKRRKNVDMWRKFDDDKVSEVPW 174
HYV W + SG W ++DDD S
Sbjct 391 HYVAWVK------------------------QESGK----------WIQYDDDNPSMQRE 416
Query 175 DSIDLAGGRSDYHVAYLLLMK 195
+ I G D+H+AY+ + K
Sbjct 417 EDITKLSGGGDWHMAYITMYK 437
> cel:C13B4.2 usp-14; Ubiquitin Specific Protease family member
(usp-14); K11843 ubiquitin carboxyl-terminal hydrolase 14
[EC:3.1.2.15]
Length=489
Score = 53.1 bits (126), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 29/99 (29%), Positives = 42/99 (42%), Gaps = 33/99 (33%)
Query 95 TGTYQLLSVVTHQGRYADSGHYVGWARAGDAGTLQGPPTAQSEDKEEEEPAAPSGPAAKR 154
+G Y L ++TH+GR + GHYV W R+ + G
Sbjct 389 SGFYDLKGIITHKGRSSQDGHYVAWMRSSEDGK--------------------------- 421
Query 155 RKNVDMWRKFDDDKVSEVPWDSIDLAGGRSDYHVAYLLL 193
WR FDD+ V+ V ++I G D+H AY+LL
Sbjct 422 ------WRLFDDEHVTVVDEEAILKTSGGGDWHSAYVLL 454
> ath:AT3G21280 UBP7; UBP7 (UBIQUITIN-SPECIFIC PROTEASE 7); ubiquitin
thiolesterase/ ubiquitin-specific protease; K11843 ubiquitin
carboxyl-terminal hydrolase 14 [EC:3.1.2.15]
Length=532
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 35/109 (32%), Positives = 51/109 (46%), Gaps = 36/109 (33%)
Query 92 PCPTGTYQLLSVVTHQGRYADSGHYVGWARAGDAGTLQGPPTAQSEDKEEEEPAAPSGPA 151
P TG Y L+SV+TH+GR ADSGHYV W + ++G
Sbjct 457 PHMTGIYDLVSVLTHKGRSADSGHYVAWVKQ-ESGK------------------------ 491
Query 152 AKRRKNVDMWRKFDDDKVS-EVPWDSIDLAGGRSDYHVAYLLLMKHILV 199
W ++DD S + D I L+GG D+H+AY+++ K L+
Sbjct 492 ---------WVQYDDANTSLQRGEDIIKLSGG-GDWHMAYIVMYKARLI 530
> hsa:84640 USP38, FLJ35970, HP43.8KD, KIAA1891; ubiquitin specific
peptidase 38 (EC:3.4.19.12); K11854 ubiquitin carboxyl-terminal
hydrolase 35/38 [EC:3.1.2.15]
Length=1042
Score = 38.1 bits (87), Expect = 0.031, Method: Composition-based stats.
Identities = 35/128 (27%), Positives = 53/128 (41%), Gaps = 20/128 (15%)
Query 88 LAGKPCPTGT----------YQLLSVVTHQGRYADSGHYVGWAR--AGDAGTLQ------ 129
LA K P+GT Y L SVV H G ++SGHY +AR + Q
Sbjct 820 LAKKLKPSGTDEASCTKLVPYLLSSVVVHSGISSESGHYYSYARNITSTDSSYQMYHQSE 879
Query 130 --GPPTAQSEDKEEEEPAAPSGPAAKRRKNVDMWRKFDDDKVSEVPWDSIDLAGGRSDYH 187
++QS + P+A + ++ W F+D +V+ + S+ R
Sbjct 880 ALALASSQSHLLGRDSPSAVFEQDLENKEMSKEWFLFNDSRVTFTSFQSVQKITSRFPKD 939
Query 188 VAYLLLMK 195
AY+LL K
Sbjct 940 TAYVLLYK 947
> tgo:TGME49_077890 hypothetical protein
Length=2530
Score = 37.7 bits (86), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 29/103 (28%), Positives = 45/103 (43%), Gaps = 22/103 (21%)
Query 97 TYQLLSVVTHQGRYADSGHYVGWARAGDAGTLQGPPTAQSEDKEEEEPAAPSGPAAKRRK 156
TY+L + V H G A+SGHY R + P ++K E +A
Sbjct 2422 TYELYAAVIHAGASANSGHYYCLGRRSEF-----PRNGAGDNKGESRRSA---------- 2466
Query 157 NVDMWRKFDDDKVSEVPWDSID--LAGGRSDYHVAYLLLMKHI 197
W KFDD +V+ V ++++ A RSD Y+L + +
Sbjct 2467 ----WFKFDDSRVTPVDEETVNEISADDRSD-DSPYMLFYRCV 2504
> sce:YNL186W UBP10, DOT4; Ubiquitin-specific protease that deubiquitinates
ubiquitin-protein moieties; may regulate silencing
by acting on Sir4p; involved in posttranscriptionally regulating
Gap1p and possibly other transporters; primarily located
in the nucleus (EC:3.1.2.15); K11873 ubiquitin carboxyl-terminal
hydrolase 10 [EC:3.1.2.15]
Length=792
Score = 35.0 bits (79), Expect = 0.21, Method: Composition-based stats.
Identities = 15/28 (53%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 98 YQLLSVVTHQGRYADSGHYVGWARAGDA 125
YQLLSVV H+GR SGHY+ + D
Sbjct 674 YQLLSVVVHEGRSLSSGHYIAHCKQPDG 701
> dre:322007 usp1, wu:fb43a02, zgc:55962; ubiquitin specific protease
1 (EC:3.4.19.12); K11832 ubiquitin carboxyl-terminal
hydrolase 1 [EC:3.1.2.15]
Length=772
Score = 34.7 bits (78), Expect = 0.29, Method: Composition-based stats.
Identities = 24/75 (32%), Positives = 34/75 (45%), Gaps = 9/75 (12%)
Query 98 YQLLSVVTHQGRYADSGHYVGWARAGDAGTLQGPPTAQSE--DKEEEEPAAPSGPAAKRR 155
Y+L +VV H G SGHY + R D P Q E D+++EE K++
Sbjct 561 YELFAVVMHSGVTISSGHYTTYIRMMDLHHTNIKPQTQDEEHDRDQEED-------MKQK 613
Query 156 KNVDMWRKFDDDKVS 170
K +DD +VS
Sbjct 614 KEETSQTDYDDGEVS 628
> mmu:245038 Dclk3, BC056929, C730036H08, Dcamkl3; doublecortin-like
kinase 3 (EC:2.7.11.1)
Length=790
Score = 33.9 bits (76), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 27/84 (32%), Positives = 35/84 (41%), Gaps = 14/84 (16%)
Query 21 AEKTTNGTASTESGAPAADGAAAATAAPEAAAVPAATNGSGAAH---SASAPAAEAAEAE 77
A+K G +E G P + GAA T+ SG H + E E
Sbjct 258 AQKWVRGKQESEPGGPPSPGAATQ----------EETHASGEKHLGVEIEKTSGEIVRCE 307
Query 78 AKKKETEL-LRLAGKPCPTGTYQL 100
K+E EL L L +PCP GT +L
Sbjct 308 KCKRERELQLGLQREPCPLGTSEL 331
> dre:553518 Phf3; si:dkey-188i13.1
Length=1445
Score = 31.6 bits (70), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 28/106 (26%), Positives = 48/106 (45%), Gaps = 8/106 (7%)
Query 9 ALRREREASSGGAEKTTNGTASTESGAPAADGAAAATAAPE-AAAVP------AATNGSG 61
L+ +A+S + + + TE+ P+ D AT +P+ A+ P
Sbjct 1308 VLKLPEKAASCSNDNMKSDSTKTEAKTPSVDSGNNATVSPKPIASAPRLDRFVIKKKEPK 1367
Query 62 AAHSASAPAAEAAEAEAKKKETE-LLRLAGKPCPTGTYQLLSVVTH 106
A + AP + + E ++KKET +L L+ KP T LS +T+
Sbjct 1368 AVKTEQAPVSVSLEISSEKKETAVVLSLSDKPADVSTESFLSSLTN 1413
> tgo:TGME49_032970 hypothetical protein
Length=1537
Score = 30.8 bits (68), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 24/61 (39%), Positives = 37/61 (60%), Gaps = 0/61 (0%)
Query 28 TASTESGAPAADGAAAATAAPEAAAVPAATNGSGAAHSASAPAAEAAEAEAKKKETELLR 87
T+ST+S A+ A+ +T P ++ V + S A+ AS+ A E+ EAE K+K+T L R
Sbjct 696 TSSTDSTGSASPRASDSTGRPLSSDVSPSLQTSSASPFASSVALESLEAELKEKKTLLDR 755
Query 88 L 88
L
Sbjct 756 L 756
> tgo:TGME49_010780 ubiquitin carboxyl-terminal hydrolase, putative
(EC:3.1.2.15)
Length=4302
Score = 29.6 bits (65), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 16/61 (26%), Positives = 30/61 (49%), Gaps = 10/61 (16%)
Query 66 ASAPAAEAAEAEAKKKETELLRLAGKPCPTGTYQLLSVVTHQGRYADSGHYVGWARAGDA 125
AS + A +++ + E+ L R Y L++++ H+G D GHYV + + +
Sbjct 1591 ASGDSRSACKSDTRDPESSLYR----------YCLVALIEHEGPATDQGHYVCYVKHLET 1640
Query 126 G 126
G
Sbjct 1641 G 1641
Lambda K H
0.310 0.125 0.356
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8145948148
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40