bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0262_orf1
Length=238
Score E
Sequences producing significant alignments: (Bits) Value
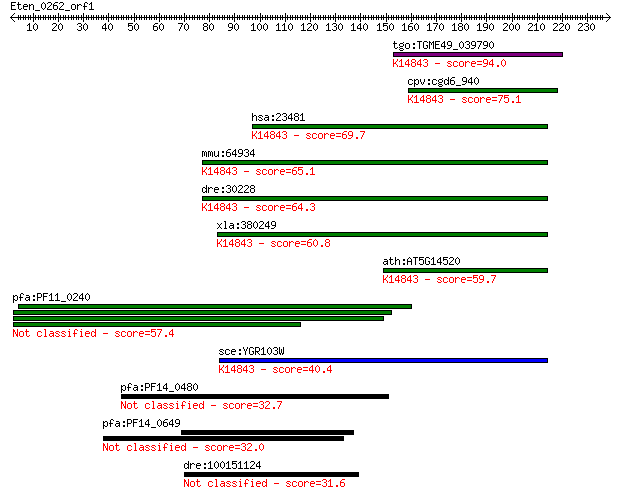
tgo:TGME49_039790 pescadillo family protein ; K14843 pescadillo 94.0 5e-19
cpv:cgd6_940 pescadillo containing BRCT domain ; K14843 pescad... 75.1 2e-13
hsa:23481 PES1, PES; pescadillo homolog 1, containing BRCT dom... 69.7 8e-12
mmu:64934 Pes1; pescadillo homolog 1, containing BRCT domain (... 65.1 2e-10
dre:30228 pes; pescadillo; K14843 pescadillo 64.3 4e-10
xla:380249 pes1, MGC53969, pes; pescadillo homolog 1, containi... 60.8 4e-09
ath:AT5G14520 pescadillo-related; K14843 pescadillo 59.7 9e-09
pfa:PF11_0240 dynein heavy chain, putative 57.4 4e-08
sce:YGR103W NOP7, YPH1; Component of several different pre-rib... 40.4 0.006
pfa:PF14_0480 conserved Plasmodium protein, unknown function 32.7 1.1
pfa:PF14_0649 conserved Plasmodium protein, unknown function 32.0 1.8
dre:100151124 retinitis pigmentosa RP1 protein-like 31.6 2.5
> tgo:TGME49_039790 pescadillo family protein ; K14843 pescadillo
Length=733
Score = 94.0 bits (232), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 41/67 (61%), Positives = 42/67 (62%), Gaps = 0/67 (0%)
Query 153 CEPHPVQNLFKGLVFFLCRETPLLPLCFAIRSCGGAVGWQSEGSPFELGDNRITHQVVDR 212
PHPVQ LF G V FL RE PLLP F IRSCGG VGWQ SPF ITH VVDR
Sbjct 455 VHPHPVQQLFDGCVIFLGREVPLLPFSFMIRSCGGKVGWQGPESPFSEDHADITHHVVDR 514
Query 213 PPEYVLQ 219
P E L+
Sbjct 515 PLECFLR 521
> cpv:cgd6_940 pescadillo containing BRCT domain ; K14843 pescadillo
Length=548
Score = 75.1 bits (183), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 29/59 (49%), Positives = 41/59 (69%), Gaps = 0/59 (0%)
Query 159 QNLFKGLVFFLCRETPLLPLCFAIRSCGGAVGWQSEGSPFELGDNRITHQVVDRPPEYV 217
+ +F+GL FF+ RE PLLPL F I+S GG VGW+++ S + D ITH V+DRP ++
Sbjct 284 KRIFEGLTFFISREVPLLPLAFVIKSFGGKVGWENQFSSIKQDDEGITHYVIDRPINFL 342
> hsa:23481 PES1, PES; pescadillo homolog 1, containing BRCT domain
(zebrafish); K14843 pescadillo
Length=588
Score = 69.7 bits (169), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 51/124 (41%), Positives = 67/124 (54%), Gaps = 9/124 (7%)
Query 97 AAEDESAAEDESAAEDESAADSSSSED--EDTEEEAEEEAAEDKAEEAAKE--TESEVEI 152
A E A + ES E +A +S + TEEEAE + E +A+E E+E
Sbjct 260 AGEGTYALDSESCMEKLAALSASLARVVVPATEEEAEVDEFPTDGEMSAQEEDRRKELEA 319
Query 153 CEPHPVQNLFKGLVFFLCRETPLLPLCFAIRSCGGAVGWQSE---GSPFELGDNRITHQV 209
E H + LF+GL FFL RE P L F IRS GG V W G+ +++ D+RITHQ+
Sbjct 320 QEKH--KKLFEGLKFFLNREVPREALAFIIRSFGGEVSWDKSLCIGATYDVTDSRITHQI 377
Query 210 VDRP 213
VDRP
Sbjct 378 VDRP 381
> mmu:64934 Pes1; pescadillo homolog 1, containing BRCT domain
(zebrafish); K14843 pescadillo
Length=584
Score = 65.1 bits (157), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 50/140 (35%), Positives = 72/140 (51%), Gaps = 19/140 (13%)
Query 77 EAAAEDDSAAEDDSAAEDESAAEDESAAEDESAAEDESAADSSSSEDEDTEEEAEEEAAE 136
E +D+ A D ++ ++ AA S A A +E+ AD ++ E T +E
Sbjct 257 ETKISEDTYALDSESSMEKLAALSASLARVVVPAIEEAEADEFPTDGEVTAQE------- 309
Query 137 DKAEEAAKETESEVEICEPHPVQNLFKGLVFFLCRETPLLPLCFAIRSCGGAVGWQSE-- 194
E+ KE E++ E H + LF+GL FFL RE P L F IRS GG V W
Sbjct 310 ---EDRKKELEAQ----EKH--KKLFEGLKFFLNREVPREALAFIIRSFGGDVSWDKSLC 360
Query 195 -GSPFELGDNRITHQVVDRP 213
G+ +++ D+ ITHQ+VDRP
Sbjct 361 IGATYDVTDSCITHQIVDRP 380
> dre:30228 pes; pescadillo; K14843 pescadillo
Length=582
Score = 64.3 bits (155), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 50/140 (35%), Positives = 62/140 (44%), Gaps = 19/140 (13%)
Query 77 EAAAEDDSAAEDDSAAEDESAAEDESAAEDESAAEDESAADSSSSEDEDTEEEAEEEAAE 136
+A E+D A E +S E SA A S E+E+ D +E ED E+ E E
Sbjct 259 KAEFEEDYALESESYTEKLSALSASLARMVASVEEEEAELDHFPTEGEDQEKMEVREKME 318
Query 137 DKAEEAAKETESEVEICEPHPVQNLFKGLVFFLCRETPLLPLCFAIRSCGGAVGWQSE-- 194
+ + K LF+GL FFL RE P L F IR GG V W
Sbjct 319 QQQSKQKK----------------LFEGLKFFLNREVPRESLAFVIRCFGGEVSWDKSLC 362
Query 195 -GSPFELGDNRITHQVVDRP 213
GS +E D ITH +VDRP
Sbjct 363 IGSTYEATDETITHHIVDRP 382
> xla:380249 pes1, MGC53969, pes; pescadillo homolog 1, containing
BRCT domain; K14843 pescadillo
Length=574
Score = 60.8 bits (146), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 51/136 (37%), Positives = 67/136 (49%), Gaps = 9/136 (6%)
Query 83 DSAAEDDSAAEDESAAEDESAAEDESAAEDESAADSSSSEDEDTEEEAEEEAAEDKA--E 140
DS +E D +S ED+ A E E E +A +S S +E + E E A E
Sbjct 251 DSFSEVDL----KSDGEDKYALETEVYMEKLAALSASLSRVIPSEPNDDTEVDEFPADPE 306
Query 141 EAAKETESEVEICEPHPVQNLFKGLVFFLCRETPLLPLCFAIRSCGGAVGWQSE---GSP 197
A E E + ++ E ++LF GL FFL RE P L F IRS GG V W + G+
Sbjct 307 NAGLEEEQKRQLQEEEKHKSLFVGLKFFLNREVPRDALAFIIRSFGGEVSWDASVCIGAT 366
Query 198 FELGDNRITHQVVDRP 213
+ D ITH +VDRP
Sbjct 367 YNSTDPSITHHIVDRP 382
> ath:AT5G14520 pescadillo-related; K14843 pescadillo
Length=590
Score = 59.7 bits (143), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 28/65 (43%), Positives = 38/65 (58%), Gaps = 5/65 (7%)
Query 149 EVEICEPHPVQNLFKGLVFFLCRETPLLPLCFAIRSCGGAVGWQSEGSPFELGDNRITHQ 208
E +C ++LFK L FFL RE P L I + GG V W+ EG+PF+ D ITH
Sbjct 329 ETRVC-----KSLFKDLKFFLSREVPRESLQLVITAFGGMVSWEGEGAPFKEDDESITHH 383
Query 209 VVDRP 213
++D+P
Sbjct 384 IIDKP 388
> pfa:PF11_0240 dynein heavy chain, putative
Length=5251
Score = 57.4 bits (137), Expect = 4e-08, Method: Composition-based stats.
Identities = 45/160 (28%), Positives = 74/160 (46%), Gaps = 8/160 (5%)
Query 4 DEEDHEATSVGSSNSSSSSSSSGSEAEETEEEIEEETKKQKNAKENKKVSSKKKERAAVA 63
+ ED +A + + + + ++ E + E+ E+E +KN ++ K V ++ AV
Sbjct 4104 NTEDEKAIEINTEDEKAVEKNTEDE-KAVEKNTEDEKAIEKNTEDEKAVEKNTEDEKAVE 4162
Query 64 AAAAAEE--EDDDDDEAAAEDDSAAEDDSAAEDESAAED--ESAAEDESAAEDESAADSS 119
E+ E + +DE A E ++ ED+ A E + E E EDE A E ++ D
Sbjct 4163 KNTEDEKAIEKNTEDEKAVEKNT--EDEKAVEQNTEDEKVVEENTEDEKAVE-KNTEDEK 4219
Query 120 SSEDEDTEEEAEEEAAEDKAEEAAKETESEVEICEPHPVQ 159
+ E +E+ EE ED+ E K E V I E VQ
Sbjct 4220 AVEKNTEDEKVVEEKIEDEKGEEQKAEEENVGIEEVEKVQ 4259
Score = 47.4 bits (111), Expect = 5e-05, Method: Composition-based stats.
Identities = 39/155 (25%), Positives = 72/155 (46%), Gaps = 25/155 (16%)
Query 2 EEDEEDHEATSVGSSNSSSSSSSSGSEAE-----ETEEEIEEETKKQKNAKENKKVSSKK 56
E++ ED +A + + + ++ E E E+ +E KN ++ K +
Sbjct 4122 EKNTEDEKAVEKNTEDEKAIEKNTEDEKAVEKNTEDEKAVE------KNTEDEKAIEKNT 4175
Query 57 KERAAVAAAAAAEEEDDDDDEAAAEDDSAAEDDSAAEDESAAEDESAAEDESAAEDESAA 116
++ AV E + +DE A E ++ ED+ E+ + EDE A E + EDE A
Sbjct 4176 EDEKAV--------EKNTEDEKAVEQNT--EDEKVVEENT--EDEKAVEKNT--EDEKAV 4221
Query 117 DSSSSEDEDTEEEAEEEAAEDKAEEAAKETESEVE 151
+ ++ +++ EE+ E+E E++ E EVE
Sbjct 4222 EKNTEDEKVVEEKIEDEKGEEQKAEEENVGIEEVE 4256
Score = 44.3 bits (103), Expect = 4e-04, Method: Composition-based stats.
Identities = 37/148 (25%), Positives = 69/148 (46%), Gaps = 16/148 (10%)
Query 2 EEDEEDHEATSVGSSNSSSSSSSSGSEAEETEEEIEEETKKQKNAKENKKVSSKKKERAA 61
E++ ED +A + + + ++ E + E+ E+E +KN ++ K V ++
Sbjct 4142 EKNTEDEKAVEKNTEDEKAVEKNTEDE-KAIEKNTEDEKAVEKNTEDEKAVEQNTEDEKV 4200
Query 62 VAAAAAAEEEDDDDDEAAAEDDSAAEDDSAAEDESAAEDESAAEDESAAEDESAADSSSS 121
V E++ +DE A E ++ ED+ A E + EDE E++ EDE + +
Sbjct 4201 V--------EENTEDEKAVEKNT--EDEKAVEKNT--EDEKVVEEK--IEDEKGEEQKAE 4246
Query 122 EDEDTEEEAEEEAAED-KAEEAAKETES 148
E+ EE E+ +D K E E ++
Sbjct 4247 EENVGIEEVEKVQIDDYKVSEKKGENKN 4274
Score = 31.6 bits (70), Expect = 2.6, Method: Composition-based stats.
Identities = 21/114 (18%), Positives = 51/114 (44%), Gaps = 4/114 (3%)
Query 2 EEDEEDHEATSVGSSNSSSSSSSSGSEAEETEEEIEEETKKQKNAKENKKVSSKKKERAA 61
E++ ED +A + + + ++ E + EE E+E +KN ++ K V ++
Sbjct 4172 EKNTEDEKAVEKNTEDEKAVEQNTEDE-KVVEENTEDEKAVEKNTEDEKAVEKNTEDEKV 4230
Query 62 VAAAAAAEEEDDDDDEAAAEDDSAAEDDSAAEDESAAEDESAAEDESAAEDESA 115
V E++ +++ A E++ E+ + + E E+++ +E+
Sbjct 4231 VEEKI---EDEKGEEQKAEEENVGIEEVEKVQIDDYKVSEKKGENKNCTYEENG 4281
> sce:YGR103W NOP7, YPH1; Component of several different pre-ribosomal
particles; forms a complex with Ytm1p and Erb1p that
is required for maturation of the large ribosomal subunit;
required for exit from G0 and the initiation of cell proliferation;
K14843 pescadillo
Length=605
Score = 40.4 bits (93), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 44/150 (29%), Positives = 76/150 (50%), Gaps = 20/150 (13%)
Query 84 SAAEDDSAAEDESAAEDESAAEDESAAEDESAADSSSSEDEDTEEEAEEEAAEDKAEEAA 143
S ED D + E++ E A+ +SA ++ + ++TE+E E+E ++K +E
Sbjct 266 SRQEDSLLKLDPTEIEEDVKVESLDASTLKSALNADEANTDETEKEEEQEKKQEKEQEKE 325
Query 144 KETESEVE------------ICEP----HPVQNLFKGLVFFLCRETPLLPLCFAIRSCGG 187
+ E+E++ + +P PV +LF VF++ RE P+ L F I SCGG
Sbjct 326 QNEETELDTFEDNNKNKGDILIQPSKYDSPVASLFSAFVFYVSREVPIDILEFLILSCGG 385
Query 188 AVGWQSEGSPFE----LGDNRITHQVVDRP 213
V ++ E + +++THQ+VDRP
Sbjct 386 NVISEAAMDQIENKKDIDMSKVTHQIVDRP 415
> pfa:PF14_0480 conserved Plasmodium protein, unknown function
Length=1653
Score = 32.7 bits (73), Expect = 1.1, Method: Composition-based stats.
Identities = 28/109 (25%), Positives = 44/109 (40%), Gaps = 12/109 (11%)
Query 45 NAKENKKVSSKKKERAAVAAAAAAEEEDDDDDEAAAEDDSAAEDDSAAEDESAAEDESAA 104
N + N S +K++ + DD E + +D + EDDS ED+S
Sbjct 976 NKRNNYYESQEKEDTWKNQKHKGINKHKDDSHEDDSHEDDSHEDDSH-------EDDSHE 1028
Query 105 EDESAAEDESAADSSSSEDEDTEEEA---EEEAAEDKAEEAAKETESEV 150
D + ED+S D S D E+E +E E ++ K E+
Sbjct 1029 ND--SHEDDSHEDDSHENDSHEEKEQYIYNKELYEKIKKKPGKNMNKEI 1075
> pfa:PF14_0649 conserved Plasmodium protein, unknown function
Length=2558
Score = 32.0 bits (71), Expect = 1.8, Method: Composition-based stats.
Identities = 20/71 (28%), Positives = 40/71 (56%), Gaps = 6/71 (8%)
Query 69 EEEDDDDDEAAAEDDSAAEDDSAAEDESAAEDESAAED---ESAAEDESAADSSSSEDED 125
E +D++ DE E+ +D+S E+++ +D++ E+ E + E+E + S E+
Sbjct 2055 ENQDENQDENQDENRDENQDESQNENQNENQDDNENEEKPNEGSNENEEKPNDGSIEN-- 2112
Query 126 TEEEAEEEAAE 136
EEA++EA +
Sbjct 2113 -NEEADKEAKD 2122
Score = 30.0 bits (66), Expect = 7.4, Method: Composition-based stats.
Identities = 18/95 (18%), Positives = 44/95 (46%), Gaps = 13/95 (13%)
Query 38 EETKKQKNAKENKKVSSKKKERAAVAAAAAAEEEDDDDDEAAAEDDSAAEDDSAAEDESA 97
++ + +N EN+ + E +D++ DE E+ +++++ E++
Sbjct 2041 QDENQDENQDENQDENQD-------------ENQDENQDENRDENQDESQNENQNENQDD 2087
Query 98 AEDESAAEDESAAEDESAADSSSSEDEDTEEEAEE 132
E+E + S +E D S +E+ ++EA++
Sbjct 2088 NENEEKPNEGSNENEEKPNDGSIENNEEADKEAKD 2122
> dre:100151124 retinitis pigmentosa RP1 protein-like
Length=3644
Score = 31.6 bits (70), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 35/75 (46%), Positives = 43/75 (57%), Gaps = 7/75 (9%)
Query 70 EEDDDDDEAAAED----DSAAEDDSAAEDESAAEDESAAEDESAAEDESAADSSSSEDED 125
EE D AAED D DD AEDE EDE+A E E ED+S+ D S+ED+
Sbjct 2706 EEKTSDGGQAAEDKTSKDGQTADDETAEDEHIIEDETAEEIE-KFEDKSSDDGQSAEDKT 2764
Query 126 TE--EEAEEEAAEDK 138
+E + A+EE AEDK
Sbjct 2765 SEDGQTADEETAEDK 2779
Lambda K H
0.296 0.115 0.298
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8274230796
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40