bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0266_orf2
Length=190
Score E
Sequences producing significant alignments: (Bits) Value
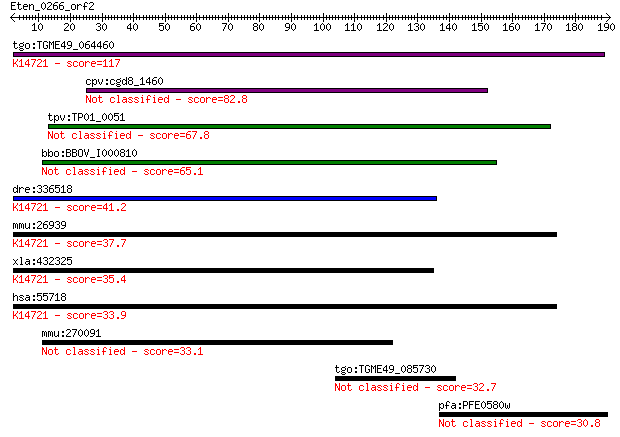
tgo:TGME49_064460 SIN-like domain-containing protein (EC:3.2.1... 117 2e-26
cpv:cgd8_1460 Sex-lethal interacting like 82.8 6e-16
tpv:TP01_0051 hypothetical protein 67.8 2e-11
bbo:BBOV_I000810 16.m00722; hypothetical protein 65.1 1e-10
dre:336518 polr3e, Sin, wu:fb12d10, wu:fs42e03, zgc:55592; pol... 41.2 0.002
mmu:26939 Polr3e, RPC5, Sin; polymerase (RNA) III (DNA directe... 37.7 0.023
xla:432325 polr3e, MGC78924; polymerase (RNA) III (DNA directe... 35.4 0.12
hsa:55718 POLR3E, RPC5, SIN; polymerase (RNA) III (DNA directe... 33.9 0.37
mmu:270091 Lrrc36, BC036564; leucine rich repeat containing 36 33.1
tgo:TGME49_085730 TBC domain-containing protein 32.7 0.74
pfa:PFE0580w kinase binding protein CGI-121, putative 30.8 2.7
> tgo:TGME49_064460 SIN-like domain-containing protein (EC:3.2.1.3);
K14721 DNA-directed RNA polymerase III subunit RPC5
Length=836
Score = 117 bits (294), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 79/218 (36%), Positives = 112/218 (51%), Gaps = 32/218 (14%)
Query 2 ATVKRLLAAAAEEQQXLQLLLQQCYLVNGNWCYKSSLACAAAYEAACRDLLLCLLVRD-- 59
AT++RL+ E + L+LLLQ LV GNW KS YE +CRDLLL LL+R
Sbjct 501 ATIQRLVTVPIAESELLRLLLQYAVLVLGNWAIKSMYVYDDMYEISCRDLLLALLIRSGA 560
Query 60 ----SQQQQQQQQQQQQQGRAGVAKEAFRAATKLPPEIADKLL---------NQVAVCVS 106
Q AG+ KEAFR+AT+L + +L +VAVC S
Sbjct 561 AFVPPSQGGAGSDACTPPASAGLPKEAFRSATRLSLDRVSHMLQEARNKTAKEEVAVCSS 620
Query 107 SSWQLKLQHDEGFAAAHPKLAAYFAAFWSRRLKEVVKEIQIKRD---------------- 150
S+W+LKL D F AAHP + +F +W +RL++VVKE+ +RD
Sbjct 621 SAWRLKLPPDAAFLAAHPAIQRHFDQWWKQRLQQVVKEVHQRRDARANPQGPLTGGEEGK 680
Query 151 SQDAAGAVASSAEVQQMGKEVQQLLTDFGGLSFEQIKK 188
++DA+ AV S A +Q G+ L G L+F+Q+++
Sbjct 681 TKDASVAV-SPALLQLAGRLCSSALKTQGALTFQQLRE 717
> cpv:cgd8_1460 Sex-lethal interacting like
Length=895
Score = 82.8 bits (203), Expect = 6e-16, Method: Composition-based stats.
Identities = 48/127 (37%), Positives = 67/127 (52%), Gaps = 15/127 (11%)
Query 25 CYLVNGNWCYKSSLACAAAYEAACRDLLLCLLVRDSQQQQQQQQQQQQQGRAGVAKEAFR 84
C LV+GNW YKS L YE+ CRDL+L LL RD AG+ +E R
Sbjct 481 CVLVSGNWVYKSELM-YNEYESCCRDLILVLLQRDEN--------------AGLNREPIR 525
Query 85 AATKLPPEIADKLLNQVAVCVSSSWQLKLQHDEGFAAAHPKLAAYFAAFWSRRLKEVVKE 144
AT LP +L +VAV S++W K D+ F + + A+Y+ +W+ R K VV+
Sbjct 526 VATDLPQIKVTNILQEVAVYRSTAWYPKFLFDKKFIEENIEEASYWKNYWAEREKYVVQY 585
Query 145 IQIKRDS 151
I+ RD+
Sbjct 586 IRDNRDT 592
> tpv:TP01_0051 hypothetical protein
Length=627
Score = 67.8 bits (164), Expect = 2e-11, Method: Composition-based stats.
Identities = 47/171 (27%), Positives = 75/171 (43%), Gaps = 26/171 (15%)
Query 13 EEQQXLQLLLQQCYLVNGNWCYKSSLACA------------AAYEAACRDLLLCLLVRDS 60
E++ ++LL + ++GNW S A Y +CR+ LLCLL R
Sbjct 381 EDEDLIRLLGKFSKNISGNWVLSSVHAIENYTEDDECNPEEMEYLVSCREFLLCLLNR-- 438
Query 61 QQQQQQQQQQQQQGRAGVAKEAFRAATKLPPEIADKLLNQVAVCVSSSWQLKLQHDEGFA 120
Q + + + +A E FR AT LP ++A+ LL VA W LKL D+ F+
Sbjct 439 --QSSEPKLKDSSSVCKLATEIFRNATGLPFKLANDLLCGVAKPQGKIWVLKLDKDDEFS 496
Query 121 AAHPKLAAYFAAFWSRRLKEVVKEIQIKRDSQDAAGAVASSAEVQQMGKEV 171
H ++ F +W RLK++ D + S ++ QM + +
Sbjct 497 EKHTQVFDLFNQYWRERLKKIA----------DIVSSYKKSPQINQMLRNI 537
> bbo:BBOV_I000810 16.m00722; hypothetical protein
Length=673
Score = 65.1 bits (157), Expect = 1e-10, Method: Composition-based stats.
Identities = 44/157 (28%), Positives = 73/157 (46%), Gaps = 18/157 (11%)
Query 11 AAEEQQXLQLLLQQCYLVNGNWCYKSSLACA------------AAYEAACRDLLLCLLVR 58
+ + + +++L + ++GNW S A AY CRD++L L R
Sbjct 402 SVTDDRLIEILQEFATNIDGNWILASDHAIPNYTIEDDYDNEDRAYLVVCRDVILGLFYR 461
Query 59 DSQQQQQQQQQQQQQGRAG-VAKEAFRAATKLPPEIADKLLNQVAVCVSSSWQLKLQHDE 117
D + Q+ G G V EA + T LP + ++L+QVA W LKL+ D+
Sbjct 462 DVVMRDLLQK-----GAYGHVTMEAVHSLTGLPVPMVHEMLSQVAQQHGKLWTLKLKRDQ 516
Query 118 GFAAAHPKLAAYFAAFWSRRLKEVVKEIQIKRDSQDA 154
F + L + A+W RL E +K+I+ R++Q +
Sbjct 517 DFYTNYNSLMKTYQAYWPNRLHECMKQIKTLRETQSS 553
> dre:336518 polr3e, Sin, wu:fb12d10, wu:fs42e03, zgc:55592; polymerase
(RNA) III (DNA directed) polypeptide E; K14721 DNA-directed
RNA polymerase III subunit RPC5
Length=696
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 37/146 (25%), Positives = 58/146 (39%), Gaps = 28/146 (19%)
Query 2 ATVKRLLAAAAEEQQXLQLLLQQCYLVNGNWCYKSSL-----ACAA----AYEAAC--RD 50
A + LLA+ + L+ + Q LV GNW KS + C++ E C RD
Sbjct 296 ANLMGLLASGTDSTAVLRCIQQVALLVQGNWVVKSDVLYPKNTCSSHSGVPAEVLCRGRD 355
Query 51 LLLCLLVRDSQQQQQQQQQQQQQGRAGVAKEAFRAATKLPPEIADKLLNQVAVC-VSSSW 109
++ + R + KE A +LPPE L Q++V ++ W
Sbjct 356 FVMWRFTIE---------------RTLMRKEV-AAIIRLPPEDVKDFLEQMSVPRLNRGW 399
Query 110 QLKLQHDEGFAAAHPKLAAYFAAFWS 135
+ L D+ F HP +A W+
Sbjct 400 EFLLPTDQDFVKKHPDVAQRQHMLWT 425
> mmu:26939 Polr3e, RPC5, Sin; polymerase (RNA) III (DNA directed)
polypeptide E; K14721 DNA-directed RNA polymerase III subunit
RPC5
Length=684
Score = 37.7 bits (86), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 46/181 (25%), Positives = 70/181 (38%), Gaps = 14/181 (7%)
Query 2 ATVKRLLAAAAEEQQXLQLLLQQCYLVNGNWCYKSSLACAAAYEAACRDLLLCLLVRDSQ 61
A + LL + + L+ + + LV GNW KS + + + +L R
Sbjct 269 ANLMSLLGPSVDSVAVLRGIQKVAMLVQGNWVVKSDILYPKDSSSPHSGMPAEVLCRGRD 328
Query 62 QQQQQQQQQQQQGRAGVAKEAFRAATKLPPEIADKLLNQVAVC-VSSSWQLKLQHDEGFA 120
+ Q + R VA A TKL E L +AV ++ W+ L +D F
Sbjct 329 FVMWKFTQSRWVVRKEVA-----AVTKLCAEDVKDFLEHMAVVRINKGWEFLLPYDLEFI 383
Query 121 AAHPKLAAYFAAFWS------RRLKEVVKEIQIKR-DSQDAA-GAVASSAEVQQMGKEVQ 172
HP + WS ++ +VKE K+ D Q A G V+ VQ + Q
Sbjct 384 KKHPDVVQRQHMLWSGIQAKLEKVYNLVKETMPKKPDGQSAPVGLVSGEQRVQTAKTKAQ 443
Query 173 Q 173
Q
Sbjct 444 Q 444
> xla:432325 polr3e, MGC78924; polymerase (RNA) III (DNA directed)
polypeptide E (80kD); K14721 DNA-directed RNA polymerase
III subunit RPC5
Length=701
Score = 35.4 bits (80), Expect = 0.12, Method: Composition-based stats.
Identities = 33/145 (22%), Positives = 53/145 (36%), Gaps = 28/145 (19%)
Query 2 ATVKRLLAAAAEEQQXLQLLLQQCYLVNGNWCYKSSLA---------CAAAYEAACR--D 50
A + L+AA E L+ + Q LV GNW KS + A E CR D
Sbjct 295 ANLMGLMAAGTESINVLRCIQQVAMLVQGNWVVKSDVVYPKDSSSPHSGVAAEVLCRGRD 354
Query 51 LLLCLLVRDSQQQQQQQQQQQQQGRAGVAKEAFRAATKLPPEIADKLLNQVAVC-VSSSW 109
++ + V ++ + TKL E L+ +++ ++ W
Sbjct 355 FVMWSFTKSRW----------------VVRKEIASVTKLCQEDVKDFLDHMSIPRINKGW 398
Query 110 QLKLQHDEGFAAAHPKLAAYFAAFW 134
+ L +DE F HP + W
Sbjct 399 EFMLPYDEDFVKKHPDVVERQNMLW 423
> hsa:55718 POLR3E, RPC5, SIN; polymerase (RNA) III (DNA directed)
polypeptide E (80kD); K14721 DNA-directed RNA polymerase
III subunit RPC5
Length=708
Score = 33.9 bits (76), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 43/181 (23%), Positives = 69/181 (38%), Gaps = 14/181 (7%)
Query 2 ATVKRLLAAAAEEQQXLQLLLQQCYLVNGNWCYKSSLACAAAYEAACRDLLLCLLVRDSQ 61
A + LL + + L+ + + LV GNW KS + + + +L R
Sbjct 295 ANLMSLLGPSIDSVAVLRGIQKVAMLVQGNWVVKSDILYPKDSSSPHSGVPAEVLCRGRD 354
Query 62 QQQQQQQQQQQQGRAGVAKEAFRAATKLPPEIADKLLNQVAVC-VSSSWQLKLQHDEGFA 120
+ Q + R VA TKL E L +AV ++ W+ L +D F
Sbjct 355 FVMWKFTQSRWVVRKEVA-----TVTKLCAEDVKDFLEHMAVVRINKGWEFILPYDGEFI 409
Query 121 AAHPKLAAYFAAFWS------RRLKEVVKEIQIKR-DSQDA-AGAVASSAEVQQMGKEVQ 172
HP + W+ ++ +VKE K+ D+Q AG V +Q + Q
Sbjct 410 KKHPDVVQRQHMLWTGIQAKLEKVYNLVKETMPKKPDAQSGPAGLVCGDQRIQVAKTKAQ 469
Query 173 Q 173
Q
Sbjct 470 Q 470
> mmu:270091 Lrrc36, BC036564; leucine rich repeat containing
36
Length=634
Score = 33.1 bits (74), Expect = 0.62, Method: Composition-based stats.
Identities = 31/114 (27%), Positives = 50/114 (43%), Gaps = 3/114 (2%)
Query 11 AAEEQQXLQLLLQQCYLVNGNWCYKSSLACAAAYEAACRDLLLCLLVRDSQQQQQQQQQQ 70
+A +L Q LV+ +W SL + RDLLL L+V QQ ++ +
Sbjct 396 SARTPHVATVLRQLLELVDKHWNGSGSLLLDKKFLGPARDLLLSLVVPAPSQQWRRSKLD 455
Query 71 QQQGRAGVAKEA-FRAATKLPPEIADKLLNQVAVCVSSSWQL--KLQHDEGFAA 121
+ G+A +E + A L P + L ++ + + L K+Q EG AA
Sbjct 456 DKAGKALCWRETELKEAGLLVPNDVESLKQKLVKVLEENLVLSEKIQQLEGTAA 509
> tgo:TGME49_085730 TBC domain-containing protein
Length=459
Score = 32.7 bits (73), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 24/39 (61%), Gaps = 1/39 (2%)
Query 104 CVSSSWQLKL-QHDEGFAAAHPKLAAYFAAFWSRRLKEV 141
C+ W + +H EGF++ H + A F FWS++LK++
Sbjct 376 CIIRLWDTYIAEHAEGFSSFHVYVCAVFLVFWSQQLKQM 414
> pfa:PFE0580w kinase binding protein CGI-121, putative
Length=179
Score = 30.8 bits (68), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 22/55 (40%), Positives = 25/55 (45%), Gaps = 2/55 (3%)
Query 137 RLKEVVKEIQIKRDSQDA--AGAVASSAEVQQMGKEVQQLLTDFGGLSFEQIKKK 189
+ E VK+ QIK DS G S +V K VQ DF LSF KKK
Sbjct 96 NINECVKQYQIKNDSSSVIYVGINISKDQVDMFIKSVQGDQADFNELSFLHDKKK 150
Lambda K H
0.318 0.127 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5429324208
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40