bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
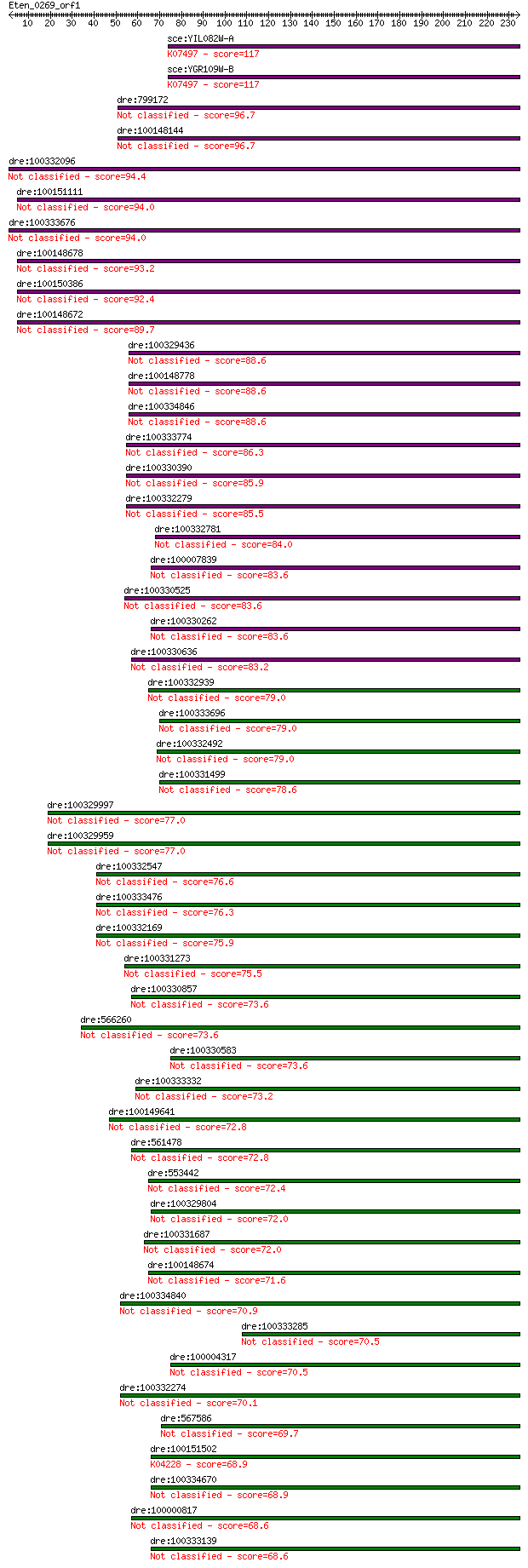
Query= Eten_0269_orf1
Length=234
Score E
Sequences producing significant alignments: (Bits) Value
sce:YIL082W-A Retrotransposon TYA Gag and TYB Pol genes; trans... 117 2e-26
sce:YGR109W-B Retrotransposon TYA Gag and TYB Pol genes; trans... 117 3e-26
dre:799172 RETRotransposon-like family member (retr-1)-like 96.7 6e-20
dre:100148144 RETRotransposon-like family member (retr-1)-like 96.7 7e-20
dre:100332096 RETRotransposon-like family member (retr-1)-like 94.4 3e-19
dre:100151111 RETRotransposon-like family member (retr-1)-like 94.0 4e-19
dre:100333676 RETRotransposon-like family member (retr-1)-like 94.0 4e-19
dre:100148678 RETRotransposon-like family member (retr-1)-like 93.2 8e-19
dre:100150386 RETRotransposon-like family member (retr-1)-like 92.4 1e-18
dre:100148672 RETRotransposon-like family member (retr-1)-like 89.7 8e-18
dre:100329436 RETRotransposon-like family member (retr-1)-like 88.6 2e-17
dre:100148778 RETRotransposon-like family member (retr-1)-like 88.6 2e-17
dre:100334846 RETRotransposon-like family member (retr-1)-like 88.6 2e-17
dre:100333774 RETRotransposon-like family member (retr-1)-like 86.3 8e-17
dre:100330390 RETRotransposon-like family member (retr-1)-like 85.9 1e-16
dre:100332279 RETRotransposon-like family member (retr-1)-like 85.5 2e-16
dre:100332781 RETRotransposon-like family member (retr-1)-like 84.0 4e-16
dre:100007839 RETRotransposon-like family member (retr-1)-like 83.6 5e-16
dre:100330525 LReO_3-like 83.6 6e-16
dre:100330262 RETRotransposon-like family member (retr-1)-like 83.6 6e-16
dre:100330636 RETRotransposon-like family member (retr-1)-like 83.2 7e-16
dre:100332939 RETRotransposon-like family member (retr-1)-like 79.0 1e-14
dre:100333696 RETRotransposon-like family member (retr-1)-like 79.0 1e-14
dre:100332492 zinc finger protein-like 79.0 1e-14
dre:100331499 RETRotransposon-like family member (retr-1)-like 78.6 2e-14
dre:100329997 RETRotransposon-like family member (retr-1)-like 77.0 6e-14
dre:100329959 RETRotransposon-like family member (retr-1)-like 77.0 6e-14
dre:100332547 RETRotransposon-like family member (retr-1)-like 76.6 7e-14
dre:100333476 RETRotransposon-like family member (retr-1)-like 76.3 8e-14
dre:100332169 LReO_3-like 75.9 1e-13
dre:100331273 RETRotransposon-like family member (retr-1)-like 75.5 2e-13
dre:100330857 LReO_3-like 73.6 6e-13
dre:566260 LReO_3-like 73.6 6e-13
dre:100330583 RETRotransposon-like family member (retr-1)-like 73.6 7e-13
dre:100333332 LReO_3-like 73.2 7e-13
dre:100149641 similar to guanylate binding protein 1, interfer... 72.8 1e-12
dre:561478 LReO_3-like 72.8 1e-12
dre:553442 ccdc33; coiled-coil domain containing 33 72.4
dre:100329804 zinc finger protein-like 72.0 2e-12
dre:100331687 retrovirus polyprotein, putative-like 72.0 2e-12
dre:100148674 Gap-Pol polyprotein-like 71.6 2e-12
dre:100334840 RETRotransposon-like family member (retr-1)-like 70.9 4e-12
dre:100333285 hypothetical protein LOC100333285 70.5 5e-12
dre:100004317 RETRotransposon-like family member (retr-1)-like 70.5 5e-12
dre:100332274 RETRotransposon-like family member (retr-1)-like 70.1 6e-12
dre:567586 LReO_3-like 69.7 9e-12
dre:100151502 retrovirus polyprotein, putative-like; K04228 ar... 68.9 1e-11
dre:100334670 retrovirus polyprotein, putative-like 68.9 2e-11
dre:100000817 LReO_3-like 68.6 2e-11
dre:100333139 retrovirus polyprotein, putative-like 68.6 2e-11
> sce:YIL082W-A Retrotransposon TYA Gag and TYB Pol genes; transcribed/translated
as one unit; polyprotein is processed to
make a nucleocapsid-like protein (Gag), reverse transcriptase
(RT), protease (PR), and integrase (IN); similar to retroviral
genes (EC:3.4.23.- 3.1.26.4 2.7.7.7 2.7.7.49); K07497
putative transposase
Length=1498
Score = 117 bits (294), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 61/162 (37%), Positives = 89/162 (54%), Gaps = 1/162 (0%)
Query 74 HHDHVT-AGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQKPAGLLEQL 132
+HDH GH G T A +S YYWP ++ Y+ +C C+ KS + GLL+ L
Sbjct 1129 YHDHTLFGGHFGVTVTLAKISPIYYWPKLQHSIIQYIRTCVQCQLIKSHRPRLHGLLQPL 1188
Query 133 LIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFTAADTVELLADR 192
I R +S+DF+T LP T+ + ILV+V+ SK A F+ +K+ A ++LL
Sbjct 1189 PIAEGRWLDISMDFVTGLPPTSNNLNMILVVVDRFSKRAHFIATRKTLDATQLIDLLFRY 1248
Query 193 LIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
+ YH FP+ + SDRD R +D + +L R IK MSS+ H
Sbjct 1249 IFSYHGFPRTITSDRDVRMTADKYQELTKRLGIKSTMSSANH 1290
> sce:YGR109W-B Retrotransposon TYA Gag and TYB Pol genes; transcribed/translated
as one unit; polyprotein is processed to
make a nucleocapsid-like protein (Gag), reverse transcriptase
(RT), protease (PR), and integrase (IN); similar to retroviral
genes (EC:3.4.23.- 3.1.26.4 2.7.7.7 2.7.7.49); K07497
putative transposase
Length=1547
Score = 117 bits (294), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 61/162 (37%), Positives = 89/162 (54%), Gaps = 1/162 (0%)
Query 74 HHDHVT-AGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQKPAGLLEQL 132
+HDH GH G T A +S YYWP ++ Y+ +C C+ KS + GLL+ L
Sbjct 1103 YHDHTLFGGHFGVTVTLAKISPIYYWPKLQHSIIQYIRTCVQCQLIKSHRPRLHGLLQPL 1162
Query 133 LIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFTAADTVELLADR 192
I R +S+DF+T LP T+ + ILV+V+ SK A F+ +K+ A ++LL
Sbjct 1163 PIAEGRWLDISMDFVTGLPPTSNNLNMILVVVDRFSKRAHFIATRKTLDATQLIDLLFRY 1222
Query 193 LIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
+ YH FP+ + SDRD R +D + +L R IK MSS+ H
Sbjct 1223 IFSYHGFPRTITSDRDVRMTADKYQELTKRLGIKSTMSSANH 1264
> dre:799172 RETRotransposon-like family member (retr-1)-like
Length=1146
Score = 96.7 bits (239), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 59/195 (30%), Positives = 96/195 (49%), Gaps = 13/195 (6%)
Query 51 RVHGLWRICVP----------QFPEFLTQT-LHSHHDHVTAGHQGQKKTFAALSEHYYWP 99
++ G+W VP PE TQ LH HD+ AGH GQ KT + E +WP
Sbjct 796 KIQGIWYRKVPLKGEGNKYQLVVPEEFTQNFLHYFHDNPLAGHLGQLKTLLKILEVAWWP 855
Query 100 GMRAYTTAYVESCTHCRASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDS 159
+R +YV+SC C+ K N KP+GLL+ LI + D + P++ +
Sbjct 856 SIRKEVWSYVKSCKLCQQYKPSNSKPSGLLQSNLITEPGHT-LGTDLMGPFPMSKKRNAY 914
Query 160 ILVMVESLSKMADFVRAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQL 219
ILV+VE +K + + S T ++L + + P+ L+SDR P+F S + + +
Sbjct 915 ILVIVEYFTKWTELFPLRDSKTQK-IAKILKEEIFTRWGVPKYLVSDRGPQFTSSILSDV 973
Query 220 CHRFNIKRCMSSSYH 234
C + + ++++YH
Sbjct 974 CKSWGCIQKLTTAYH 988
> dre:100148144 RETRotransposon-like family member (retr-1)-like
Length=1272
Score = 96.7 bits (239), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 59/195 (30%), Positives = 97/195 (49%), Gaps = 13/195 (6%)
Query 51 RVHGLWRICVP----------QFPEFLTQT-LHSHHDHVTAGHQGQKKTFAALSEHYYWP 99
++ G+W VP PE LTQ LH HD+ AGH GQ KT + E +WP
Sbjct 939 KIQGIWYRKVPLKGEGNKYQLVVPEELTQNFLHYFHDNPLAGHLGQLKTLLKILEVAWWP 998
Query 100 GMRAYTTAYVESCTHCRASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDS 159
+R +YV+SC C+ K N KP+GLL+ LI + D + P++ +
Sbjct 999 SVRKEVWSYVKSCKLCQQYKPSNSKPSGLLQSNLITEPGHT-LGTDLMGPFPMSKKRNAY 1057
Query 160 ILVMVESLSKMADFVRAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQL 219
ILV+V+ +K + + S T ++L + + P+ L+SDR P+F S + + +
Sbjct 1058 ILVIVDYFTKWTELFPLRDSKTQK-IAKILKEEIFTRWGVPKYLVSDRGPQFTSSILSDV 1116
Query 220 CHRFNIKRCMSSSYH 234
C + + ++++YH
Sbjct 1117 CKSWGCIQKLTTAYH 1131
> dre:100332096 RETRotransposon-like family member (retr-1)-like
Length=456
Score = 94.4 bits (233), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 60/234 (25%), Positives = 112/234 (47%), Gaps = 8/234 (3%)
Query 1 PSCQWPEAYSTCPVFRVPYKAAANQPREDLQIEFCNRQFTFRYLQPYLHIRVHGLWRICV 60
P W E + + ++ A N+ +E Q ++ + ++ VH +++ +
Sbjct 23 PQTIWEEQHQDTDIMKIFQALAKNEQQE--QAQYTVLEDKLYHITHLADETVH--YKVVI 78
Query 61 PQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKS 120
P L +HD +GH G KT+ + + YWPGM YV++C C+ +K
Sbjct 79 PS--TLRPTVLEWYHDTPLSGHLGIYKTYKRIQDVAYWPGMWTDIKKYVKNCAKCQVTKW 136
Query 121 LNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSF 180
N+KPAG L+Q+ SR +D + +P + ++ +LV V+ SK + + +
Sbjct 137 DNRKPAGKLQQVTT-SRPNEMWGVDIMGPMPKSGKQNEYLLVFVDYFSKWVELFPMRHA- 194
Query 181 TAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
TA +L ++ P ++SDR +F S L+ +LC ++NI ++++YH
Sbjct 195 TAQTIATILRQEMLTRWGVPDFILSDRGAQFVSSLFTELCGKWNITPKLTTAYH 248
> dre:100151111 RETRotransposon-like family member (retr-1)-like
Length=1585
Score = 94.0 bits (232), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 66/231 (28%), Positives = 115/231 (49%), Gaps = 11/231 (4%)
Query 5 WPEAYSTCPVFRVPYKAAANQPREDLQIEFCNRQFTFRYLQPYL-HIRVHGLWRICVPQF 63
W E +S ++ +A A +P + Q E + Y + YL + +VH +R+ VP
Sbjct 1152 WEEQHSDEETTKL-LQAVAEEPNQLEQYEVIEDKL---YHKTYLKNDQVH--YRVYVPN- 1204
Query 64 PEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQ 123
LH +H H +GH G KT+ + +WPG+ +V+ C C+ K NQ
Sbjct 1205 -RLRPTLLHHYHSHPLSGHHGIYKTYKRIQAVAFWPGLWTDVKRHVKECVKCQTIKYDNQ 1263
Query 124 KPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFTAA 183
KPAG L Q I SR + +D I LP +T ++ +LV V+ SK +F +++ +
Sbjct 1264 KPAGKL-QSTITSRPNQMLGVDIIGPLPRSTQQNEYLLVFVDYYSKWVEFFPMRQANAQS 1322
Query 184 DTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
V + L R+ P ++SDR +F S ++ +C ++ + + ++++YH
Sbjct 1323 VAVIFRREILTRW-GVPDFILSDRGTQFISSVFKNVCEKWGVTQKLTTAYH 1372
> dre:100333676 RETRotransposon-like family member (retr-1)-like
Length=922
Score = 94.0 bits (232), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 60/234 (25%), Positives = 112/234 (47%), Gaps = 8/234 (3%)
Query 1 PSCQWPEAYSTCPVFRVPYKAAANQPREDLQIEFCNRQFTFRYLQPYLHIRVHGLWRICV 60
P W E + + ++ A N+ +E Q ++ + ++ VH +++ +
Sbjct 486 PQTIWEEQHQDTDIMKIFQALAKNEQQE--QGQYTVLEDKLYHITHLADETVH--YKVVI 541
Query 61 PQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKS 120
P L +HD +GH G KT+ + + YWPGM YV++C C+ +K
Sbjct 542 PS--TLRPTVLEWYHDTPLSGHLGIYKTYKRIQDVAYWPGMWTDIKKYVKNCAKCQVTKW 599
Query 121 LNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSF 180
N+KPAG L+Q+ SR +D + +P + ++ +LV V+ SK + + +
Sbjct 600 DNRKPAGKLQQVTT-SRPNEMWGVDIMGPMPKSGKQNEYLLVFVDYFSKWVELFPMRHA- 657
Query 181 TAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
TA +L ++ P ++SDR +F S L+ +LC ++NI ++++YH
Sbjct 658 TAQTIATILRQEMLTRWGVPDFILSDRGAQFVSSLFTELCGKWNITPKLTTAYH 711
> dre:100148678 RETRotransposon-like family member (retr-1)-like
Length=866
Score = 93.2 bits (230), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 65/231 (28%), Positives = 115/231 (49%), Gaps = 11/231 (4%)
Query 5 WPEAYSTCPVFRVPYKAAANQPREDLQIEFCNRQFTFRYLQPYL-HIRVHGLWRICVPQF 63
W E +S ++ +A A +P + Q E + Y + YL + +VH +R+ VP
Sbjct 537 WEEQHSDEETTKL-LQAVAEEPNQLEQYEVIEDKL---YHKTYLKNDQVH--YRVYVPN- 589
Query 64 PEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQ 123
LH +H H +GH G KT+ + +WPG+ +V+ C C+ K NQ
Sbjct 590 -RLRPTLLHHYHSHPLSGHHGIYKTYKRIQAVAFWPGLWTDVKRHVKECVKCQTIKYDNQ 648
Query 124 KPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFTAA 183
KPAG L Q I SR + +D + LP +T ++ +LV V+ SK +F +++ +
Sbjct 649 KPAGKL-QSTITSRPNQMLGVDIMGPLPRSTQQNEYLLVFVDYYSKWVEFFPMRQANAQS 707
Query 184 DTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
V + L R+ P ++SDR +F S ++ +C ++ + + ++++YH
Sbjct 708 VAVIFRREILTRW-GVPDFILSDRGTQFISSVFKNVCEKWGVTQKLTTAYH 757
> dre:100150386 RETRotransposon-like family member (retr-1)-like
Length=1176
Score = 92.4 bits (228), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 65/231 (28%), Positives = 115/231 (49%), Gaps = 11/231 (4%)
Query 5 WPEAYSTCPVFRVPYKAAANQPREDLQIEFCNRQFTFRYLQPYL-HIRVHGLWRICVPQF 63
W E +S ++ +A A +P + Q E + Y + YL + +VH +R+ VP
Sbjct 716 WEEQHSDEETTKL-LQAVAEEPNQLEQYEVIEDKL---YHKTYLKNDQVH--YRVYVPN- 768
Query 64 PEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQ 123
LH +H H +GH G KT+ + +WPG+ +V+ C C+ K NQ
Sbjct 769 -RLRPTLLHHYHSHPLSGHHGIYKTYKRIQAVAFWPGLWTDVKRHVKECVKCQTIKYDNQ 827
Query 124 KPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFTAA 183
KPAG L Q I SR + +D + LP +T ++ +LV V+ SK +F +++ +
Sbjct 828 KPAGKL-QSTITSRPNQMLGVDIMGPLPRSTQQNEYLLVFVDYYSKWVEFFPMRQANAQS 886
Query 184 DTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
V + L R+ P ++SDR +F S ++ +C ++ + + ++++YH
Sbjct 887 VAVIFRREILTRW-GVPDFILSDRGTQFISSVFKNVCEKWGVTQKLTTAYH 936
> dre:100148672 RETRotransposon-like family member (retr-1)-like
Length=1398
Score = 89.7 bits (221), Expect = 8e-18, Method: Compositional matrix adjust.
Identities = 64/231 (27%), Positives = 114/231 (49%), Gaps = 11/231 (4%)
Query 5 WPEAYSTCPVFRVPYKAAANQPREDLQIEFCNRQFTFRYLQPYL-HIRVHGLWRICVPQF 63
W E +S ++ +A A +P + Q E + Y + YL + +VH +R+ VP
Sbjct 965 WEEQHSDEETTKL-LQAVAEEPNQLEQYEVIEDKL---YHKTYLKNDQVH--YRVYVPN- 1017
Query 64 PEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQ 123
LH +H H +GH G KT+ + +WPG+ +V+ C C+ K NQ
Sbjct 1018 -RLRPTLLHHYHSHPLSGHHGIYKTYKRIQAVAFWPGLWTDVKRHVKECVKCQTIKYDNQ 1076
Query 124 KPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFTAA 183
KPAG L Q I SR + +D + LP +T ++ + V V+ SK +F +++ +
Sbjct 1077 KPAGKL-QSTITSRPNQMLVVDIMGPLPRSTQQNEYLWVFVDYYSKWVEFFPMRQANAQS 1135
Query 184 DTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
V + L R+ P ++SDR +F S ++ +C ++ + + ++++YH
Sbjct 1136 VAVIFRREILTRW-GVPDFILSDRGTQFISSVFKNVCEKWGVTQKLTTAYH 1185
> dre:100329436 RETRotransposon-like family member (retr-1)-like
Length=1188
Score = 88.6 bits (218), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 54/180 (30%), Positives = 98/180 (54%), Gaps = 6/180 (3%)
Query 56 WRICVPQ-FPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTH 114
++I VPQ E L L ++H + +GH G+ KT L + +WP M + +V++CT
Sbjct 808 FQIYVPQTLREIL---LEAYHSNPLSGHFGRYKTQKRLMQVAFWPNMWRDVSDFVKNCTS 864
Query 115 CRASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFV 174
C+ +K +KPAG L+Q + + +D + LP +T G+ +LV+V+ S +
Sbjct 865 CQQNKPECRKPAGKLQQTEV-KEPWEMLGVDLMGPLPRSTLGNTQLLVVVDYYSHWVEMF 923
Query 175 RAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
+K+ TA + L ++ P+ L+SDR P+F S++ LC R+ + + ++++YH
Sbjct 924 PLRKA-TAGVIAQTLRKEVLTRWGVPKFLLSDRGPQFTSEILKDLCSRWGVVQKLTTAYH 982
> dre:100148778 RETRotransposon-like family member (retr-1)-like
Length=1151
Score = 88.6 bits (218), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 54/180 (30%), Positives = 98/180 (54%), Gaps = 6/180 (3%)
Query 56 WRICVPQ-FPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTH 114
++I VPQ E L L ++H + +GH G+ KT L + +WP M + +V++CT
Sbjct 763 FQIYVPQTLREIL---LEAYHSNPLSGHFGRYKTQKRLMQVAFWPNMWRDVSDFVKNCTS 819
Query 115 CRASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFV 174
C+ +K +KPAG L+Q + + +D + LP +T G+ +LV+V+ S +
Sbjct 820 CQQNKPECRKPAGKLQQTEV-KEPWEMLGVDLMGPLPRSTLGNTQLLVVVDYYSHWVEMF 878
Query 175 RAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
+K+ TA + L ++ P+ L+SDR P+F S++ LC R+ + + ++++YH
Sbjct 879 PLRKA-TAGVIAQTLRKEVLTRWGVPKFLLSDRGPQFTSEILKDLCSRWGVVQKLTTAYH 937
> dre:100334846 RETRotransposon-like family member (retr-1)-like
Length=1402
Score = 88.6 bits (218), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 54/180 (30%), Positives = 98/180 (54%), Gaps = 6/180 (3%)
Query 56 WRICVPQ-FPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTH 114
++I VPQ E L L ++H + +GH G+ KT L + +WP M + +V++CT
Sbjct 967 FQIYVPQTLREIL---LEAYHSNPLSGHFGRYKTQKRLMQVAFWPNMWRDVSDFVKNCTS 1023
Query 115 CRASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFV 174
C+ +K +KPAG L+Q + + +D + LP +T G+ +LV+V+ S +
Sbjct 1024 CQQNKPECRKPAGKLQQTEV-KEPWEMLGVDLMGPLPRSTLGNTQLLVVVDYYSHWVEMF 1082
Query 175 RAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
+K+ TA + L ++ P+ L+SDR P+F S++ LC R+ + + ++++YH
Sbjct 1083 PLRKA-TAGVIAQTLRKEVLTRWGVPKFLLSDRGPQFTSEILKDLCSRWGVVQKLTTAYH 1141
> dre:100333774 RETRotransposon-like family member (retr-1)-like
Length=1154
Score = 86.3 bits (212), Expect = 8e-17, Method: Compositional matrix adjust.
Identities = 51/181 (28%), Positives = 93/181 (51%), Gaps = 5/181 (2%)
Query 55 LWRICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTH 114
++++ VP F Q L HDHV +GH G KT+ + +H++WPG++ Y +C
Sbjct 10 VYQVVVPSV--FRQQVLVLAHDHVLSGHLGITKTYHRILKHFFWPGLKQDVARYCRTCQS 67
Query 115 CRASKSLNQK-PAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADF 173
C+ S NQ P L + I V +D + LP T G+ +L ++ + ++ +
Sbjct 68 CQFSGKPNQVIPPAPLNPIPIMMEPFERVIVDCVGPLPKTKAGNQFLLTIMCAATRFPEA 127
Query 174 VRAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSY 233
+ +K TA ++ L + P+V+ SD+ F S+++N++ +I+ C+SS+Y
Sbjct 128 IPLRK-ITAPVIIKALT-KFFATFGLPKVVQSDQGTNFMSNVFNEVLRSLSIQHCVSSAY 185
Query 234 H 234
H
Sbjct 186 H 186
> dre:100330390 RETRotransposon-like family member (retr-1)-like
Length=650
Score = 85.9 bits (211), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 52/184 (28%), Positives = 95/184 (51%), Gaps = 11/184 (5%)
Query 55 LWRICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTH 114
++++ VP F Q L HDHV +GH G KT+ + +H++WPG++ Y +C
Sbjct 10 VYQVVVPSV--FRQQVLVLAHDHVLSGHLGITKTYHRILKHFFWPGLKQDVARYCRTCQS 67
Query 115 CRASKSLNQ----KPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKM 170
C+ S NQ P + L+ P R V +D + LP T G+ +L ++ + ++
Sbjct 68 CQFSGKPNQVIPPAPLNPIPILMEPFER---VIVDCVGPLPKTKAGNQFLLTIMCAATRF 124
Query 171 ADFVRAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMS 230
+ + +K TA ++ L + P+V+ SD+ F S+++N++ +I+ C+S
Sbjct 125 PEAIPLRK-ITAPVIIKALT-KFFATFGLPKVVQSDQGTNFMSNVFNEVLRSLSIQHCVS 182
Query 231 SSYH 234
S+YH
Sbjct 183 SAYH 186
> dre:100332279 RETRotransposon-like family member (retr-1)-like
Length=1088
Score = 85.5 bits (210), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 52/184 (28%), Positives = 95/184 (51%), Gaps = 11/184 (5%)
Query 55 LWRICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTH 114
++++ VP F Q L HDHV +GH G KT+ + +H++WPG++ Y +C
Sbjct 448 VYQVVVPSV--FRQQVLVLAHDHVLSGHLGITKTYHRILKHFFWPGLKQDVARYCRTCQS 505
Query 115 CRASKSLNQ----KPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKM 170
C+ S NQ P + L+ P R V +D + LP T G+ +L ++ + ++
Sbjct 506 CQFSGKPNQVIPPAPLNPIPILMEPFER---VIVDCVGPLPKTKAGNQFLLTIMCAATRF 562
Query 171 ADFVRAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMS 230
+ + +K TA ++ L + P+V+ SD+ F S+++N++ +I+ C+S
Sbjct 563 PEAIPLRK-ITAPVIIKALT-KFFATFGLPKVVQSDQGTNFMSNVFNEVLRSLSIQHCVS 620
Query 231 SSYH 234
S+YH
Sbjct 621 SAYH 624
> dre:100332781 RETRotransposon-like family member (retr-1)-like
Length=601
Score = 84.0 bits (206), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 47/171 (27%), Positives = 87/171 (50%), Gaps = 9/171 (5%)
Query 68 TQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQK-PA 126
T L H+HV +GH G KTF +S ++YWPG+++ + + +SC C+ + NQK P
Sbjct 36 TYVLKLAHEHVMSGHLGVTKTFYRVSRYFYWPGIKSAVSEFCKSCKVCQLTGKPNQKVPV 95
Query 127 GLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFTAADTV 186
L + + + + +D + LP + +GH IL ++ + ++ + V +
Sbjct 96 APLTPIPVINEPFERLIIDCVGPLPKSKSGHQYILTIMCAATRFPEAVPMRNIKAKG--- 152
Query 187 ELLADRLIRYHS---FPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
+A LI+Y S P+++ +DR F S ++ ++ F + +SS YH
Sbjct 153 --IAKELIKYCSIFGLPKIIQTDRGTNFTSKMFEKVLKEFGVNHQLSSPYH 201
> dre:100007839 RETRotransposon-like family member (retr-1)-like
Length=731
Score = 83.6 bits (205), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 48/170 (28%), Positives = 87/170 (51%), Gaps = 3/170 (1%)
Query 66 FLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQK- 124
+ +Q L HDHV +GH G +KT+ + +++WPG+++ +AY SC C+ + NQ
Sbjct 156 YRSQILALAHDHVFSGHLGVRKTYDRILRYFFWPGLKSDVSAYCRSCHSCQLAGKPNQVI 215
Query 125 PAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFTAAD 184
P L + I S + +D + LP + GH IL ++ + ++ + V + A
Sbjct 216 PPAPLHPIPITSEPFERILIDCVGPLPKSKAGHQYILTIMCAATRYPEAVPMRNIKAKAV 275
Query 185 TVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
EL+ + P+V+ +D+ F S L++Q+ +K +SS+YH
Sbjct 276 VRELI--KFCSTFGLPKVIQTDQGSNFTSTLFSQVLRELTVKHQLSSAYH 323
> dre:100330525 LReO_3-like
Length=1366
Score = 83.6 bits (205), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 48/183 (26%), Positives = 97/183 (53%), Gaps = 7/183 (3%)
Query 54 GLWRICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCT 113
++++ +P+ ++ L HDH +GH G +KT+ L +H++WPGM++ + Y +SC
Sbjct 550 SVFQVVIPK--DYREYVLSVAHDHELSGHLGIRKTYNNLLKHFFWPGMKSTVSHYCQSCH 607
Query 114 HCRASKSLNQ--KPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMA 171
C+ + NQ PA L+ + + + + +D + LP T +GH +L ++ S ++
Sbjct 608 ACQVAGKPNQVISPAP-LKPIPVMTEPFEKLVVDCVGPLPRTKSGHSYLLTLMCSATRFP 666
Query 172 DFVRAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSS 231
+ + +S A V+ + + P+ + SD+ F S ++ ++ NI+ C+SS
Sbjct 667 EAI-PLRSLKAPTIVKAIV-KFCTTFGLPKFIQSDQGSNFMSRIFRKVMKELNIQHCVSS 724
Query 232 SYH 234
+YH
Sbjct 725 AYH 727
> dre:100330262 RETRotransposon-like family member (retr-1)-like
Length=728
Score = 83.6 bits (205), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 48/170 (28%), Positives = 87/170 (51%), Gaps = 3/170 (1%)
Query 66 FLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQK- 124
+ +Q L HDHV +GH G +KT+ + +++WPG+++ +AY SC C+ + NQ
Sbjct 156 YRSQILALAHDHVFSGHLGVRKTYDRILRYFFWPGLKSDVSAYCRSCHSCQLAGKPNQVI 215
Query 125 PAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFTAAD 184
P L + I S + +D + LP + GH IL ++ + ++ + V + A
Sbjct 216 PPAPLHPIPITSEPFERILIDCVGPLPKSKAGHQYILTIMCAATRYPEAVPMRNIKAKAV 275
Query 185 TVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
EL+ + P+V+ +D+ F S L++Q+ +K +SS+YH
Sbjct 276 VRELI--KFCSTFGLPKVIQTDQGSNFTSTLFSQVLRELTVKHQLSSAYH 323
> dre:100330636 RETRotransposon-like family member (retr-1)-like
Length=1406
Score = 83.2 bits (204), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 51/179 (28%), Positives = 91/179 (50%), Gaps = 5/179 (2%)
Query 57 RICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCR 116
+I VP + +Q L HDHV +GH G +KT+ + +++WPG+++ +AY SC C+
Sbjct 535 QIVVPV--GYRSQILALAHDHVFSGHLGVRKTYDRILRYFFWPGLKSDVSAYCRSCHSCQ 592
Query 117 ASKSLNQK-PAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVR 175
+ NQ P L + I S + +D + LP + GH IL ++ + ++ + V
Sbjct 593 LAGKPNQVIPPAPLHPIPITSEPFERILIDCVGPLPKSKAGHQYILTIMCAATRYPEAVP 652
Query 176 AKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
+ A EL+ + P+V+ +D+ F S L++Q+ +K +SS+YH
Sbjct 653 MRNIKAKAVVRELI--KFCSTFGLPKVIQTDQGSNFTSTLFSQVLRELTVKHQLSSAYH 709
> dre:100332939 RETRotransposon-like family member (retr-1)-like
Length=733
Score = 79.0 bits (193), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 47/174 (27%), Positives = 88/174 (50%), Gaps = 9/174 (5%)
Query 65 EFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQK 124
++ Q L+ HDH +GH G KT+ + +H++WPG++ ++ +C C+ + NQK
Sbjct 192 QYREQVLNLAHDHPLSGHLGVTKTYKRILKHFFWPGLKKNVVSHCRTCHVCQVTGKPNQK 251
Query 125 -PAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFTAA 183
P L + I HV LD + LP T G+ +L ++ S ++ + + +K
Sbjct 252 IPPAPLVPIPIVGEPFEHVILDCVGPLPKTKAGNQFLLTIMCSATRFPEAIPLRKI---- 307
Query 184 DTVELLADRLIRYHS---FPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
T ++ +I++ S P+++ +D+ F S ++ Q+ IK SS+YH
Sbjct 308 -TAPVVIKAMIKFFSTFGLPKIVQTDQGTNFLSKVFTQVLTSLGIKHRTSSAYH 360
> dre:100333696 RETRotransposon-like family member (retr-1)-like
Length=1036
Score = 79.0 bits (193), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 42/166 (25%), Positives = 81/166 (48%), Gaps = 3/166 (1%)
Query 70 TLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQK-PAGL 128
L H+H +GH G KTF +S +++WPG+++ + + + C C+ + NQ P+
Sbjct 299 VLQLAHEHPLSGHLGVNKTFKRISRYFFWPGLKSSVSKFCKQCHVCQIAGKPNQTIPSAP 358
Query 129 LEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFTAADTVEL 188
L + + LD + LP + +GH IL ++ + ++ + + + A E+
Sbjct 359 LHPIPAVGEPFERLILDCVGPLPKSKSGHQYILTLMCTATRFPEAIPLRSLKAKAIVKEV 418
Query 189 LADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
+ + P+V+ +DR F S L+ Q+C + +SS+YH
Sbjct 419 V--KFCSTFGLPKVIQTDRGTNFTSKLFTQICKELGVTNQLSSAYH 462
> dre:100332492 zinc finger protein-like
Length=1439
Score = 79.0 bits (193), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 53/172 (30%), Positives = 90/172 (52%), Gaps = 14/172 (8%)
Query 69 QTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQK-PAG 127
Q L HD+V AGH G KT+ + +++WPGM+A Y SC C+ NQK P
Sbjct 622 QVLSVAHDNV-AGHLGITKTYHRILRYFFWPGMKADVAKYCRSCHTCQIVGKPNQKIPPA 680
Query 128 LLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFV--RAKKSFTAADT 185
L+ + + + V LD + LP T +GH +L ++ + ++ + + R+ K+
Sbjct 681 PLKPIPVVDEPFSRVILDCVGPLPRTKSGHVYLLTLMCTTTRYPEAIPLRSLKA------ 734
Query 186 VELLADRLIRYHS---FPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
+++ LI + S FP+V+ +D+ F S ++ Q+ + NIK SS+YH
Sbjct 735 -KVILKALITFFSTFGFPKVIQTDQGTNFMSRVFKQVLSQLNIKHITSSAYH 785
> dre:100331499 RETRotransposon-like family member (retr-1)-like
Length=1264
Score = 78.6 bits (192), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 42/166 (25%), Positives = 81/166 (48%), Gaps = 3/166 (1%)
Query 70 TLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQK-PAGL 128
L H+H +GH G KTF +S +++WPG+++ + + + C C+ + NQ P+
Sbjct 444 VLQLAHEHPLSGHLGVNKTFKRISRYFFWPGLKSSVSKFCKQCHVCQIAGKPNQTIPSAP 503
Query 129 LEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFTAADTVEL 188
L + + LD + LP + +GH IL ++ + ++ + + + A E+
Sbjct 504 LHPIPAVGEPFERLILDCVGPLPKSKSGHQYILTLMCTATRFPEAIPLRSLKAKAIVKEV 563
Query 189 LADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
+ + P+V+ +DR F S L+ Q+C + +SS+YH
Sbjct 564 V--KFCSTFGLPKVIQTDRGTNFTSKLFTQICKELGVTNQLSSAYH 607
> dre:100329997 RETRotransposon-like family member (retr-1)-like
Length=915
Score = 77.0 bits (188), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 60/220 (27%), Positives = 102/220 (46%), Gaps = 14/220 (6%)
Query 19 YKAAANQPREDLQIEFCNRQFTFRYLQPYLHIRVHGLWRICVPQF--PEFL-TQTLHSHH 75
++AA + +I +C + YL RV + CV Q P L Q LH H
Sbjct 560 WEAAKQATTDSSRIAYCVQN-------DYLFRRVPNKDQGCVYQLVIPASLREQFLHFAH 612
Query 76 DHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQKPAGLLEQLLIP 135
+ +GH G+ KT L + YWP +R ++ C C+ K K +GLL+ P
Sbjct 613 SNPLSGHLGRMKTLKRLLDSVYWPEVRKDVWSFCTQCKTCQIYKPRISKLSGLLQS--TP 670
Query 136 SRRRAH-VSLDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFTAADTVELLADRLI 194
+ + +D + P + ++ +LV V+ SK + + + T T L +
Sbjct 671 VVEPGYMLGVDLMGPFPKSNRSNEFLLVFVDYCSKWVELFALRSAKTHLITNILTKEMFT 730
Query 195 RYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
R+ + P L+SDR P+F + L N+ C R+ + + +S++YH
Sbjct 731 RWGT-PAYLVSDRGPQFTAQLLNETCKRWGVVQKLSTAYH 769
> dre:100329959 RETRotransposon-like family member (retr-1)-like
Length=927
Score = 77.0 bits (188), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 60/220 (27%), Positives = 102/220 (46%), Gaps = 14/220 (6%)
Query 19 YKAAANQPREDLQIEFCNRQFTFRYLQPYLHIRVHGLWRICVPQF--PEFL-TQTLHSHH 75
++AA + +I +C + YL RV + CV Q P L Q LH H
Sbjct 560 WEAAKQATTDSSRIAYCVQN-------DYLFRRVPNKDQGCVYQLVIPASLREQFLHFAH 612
Query 76 DHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQKPAGLLEQLLIP 135
+ +GH G+ KT L + YWP +R ++ C C+ K K +GLL+ P
Sbjct 613 SNPLSGHLGRMKTLKRLLDSVYWPEIRKDVWSFCTQCKTCQIYKPRISKLSGLLQS--TP 670
Query 136 SRRRAH-VSLDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFTAADTVELLADRLI 194
+ + +D + P + ++ +LV V+ SK + + + T T L +
Sbjct 671 VVEPGYMLGVDLMGPFPKSNRSNEFLLVFVDYCSKWVELFALRSAKTHLITNILTKEMFT 730
Query 195 RYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
R+ + P L+SDR P+F + L N+ C R+ + + +S++YH
Sbjct 731 RWGT-PAYLVSDRGPQFTAQLLNETCKRWGVVQKLSTAYH 769
> dre:100332547 RETRotransposon-like family member (retr-1)-like
Length=850
Score = 76.6 bits (187), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 46/195 (23%), Positives = 93/195 (47%), Gaps = 5/195 (2%)
Query 41 FRYLQPYLHIRVHGLWRICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPG 100
R +P ++ V + ++ +P + Q L H+H AGH G KT+ + ++++WPG
Sbjct 136 MRRWKPQVNGDVLAVHQVVLPS--TYRPQVLKLAHEHPLAGHLGITKTYKRILKYFFWPG 193
Query 101 MRAYTTAYVESCTHCRASKSLNQK-PAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDS 159
++ +Y +C C+ S NQ P L+ + + + + LD + LP + +GH
Sbjct 194 LKNSVVSYCRACHDCQLSGKPNQTVPTAPLQPIPVINEPFERLILDCVGPLPKSKSGHQY 253
Query 160 ILVMVESLSKMADFVRAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQL 219
IL ++ + ++ + + + E++ + P+V+ +D+ F S L+ Q+
Sbjct 254 ILTLMCAATRFPEAFPLRSLRASVIVKEII--KFCSTFGLPKVIQTDQGSNFTSKLFAQV 311
Query 220 CHRFNIKRCMSSSYH 234
+ MSS+YH
Sbjct 312 LKELGVSHQMSSAYH 326
> dre:100333476 RETRotransposon-like family member (retr-1)-like
Length=1091
Score = 76.3 bits (186), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 46/195 (23%), Positives = 93/195 (47%), Gaps = 5/195 (2%)
Query 41 FRYLQPYLHIRVHGLWRICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPG 100
R +P ++ V + ++ +P + Q L H+H AGH G KT+ + ++++WPG
Sbjct 368 MRRWKPQVNGDVLAVHQVVLPS--AYRPQVLKLAHEHPLAGHLGITKTYKRILKYFFWPG 425
Query 101 MRAYTTAYVESCTHCRASKSLNQK-PAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDS 159
++ +Y +C C+ S NQ P L+ + + + + LD + LP + +GH
Sbjct 426 LKNSVVSYCRACHDCQLSGKPNQTVPTAPLQPIPVINEPFERLILDCVGPLPKSKSGHQY 485
Query 160 ILVMVESLSKMADFVRAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQL 219
IL ++ + ++ + + + E++ + P+V+ +D+ F S L+ Q+
Sbjct 486 ILTLMCAATRFPEAFPLRSLRASVIVKEII--KFCSTFGLPKVIQTDQGSNFTSKLFAQV 543
Query 220 CHRFNIKRCMSSSYH 234
+ MSS+YH
Sbjct 544 LKELGVSHQMSSAYH 558
> dre:100332169 LReO_3-like
Length=1775
Score = 75.9 bits (185), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 46/195 (23%), Positives = 93/195 (47%), Gaps = 5/195 (2%)
Query 41 FRYLQPYLHIRVHGLWRICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPG 100
R +P ++ V + ++ +P + Q L H+H AGH G KT+ + ++++WPG
Sbjct 884 MRRWKPQVNGDVLAVHQVVLPS--AYRPQVLKLAHEHPLAGHLGITKTYKRILKYFFWPG 941
Query 101 MRAYTTAYVESCTHCRASKSLNQK-PAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDS 159
++ +Y +C C+ S NQ P L+ + + + + LD + LP + +GH
Sbjct 942 LKNSVVSYCRACHDCQLSGKPNQTVPTAPLQPIPVINEPFERLILDCVGPLPKSKSGHQY 1001
Query 160 ILVMVESLSKMADFVRAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQL 219
IL ++ + ++ + + + E++ + P+V+ +D+ F S L+ Q+
Sbjct 1002 ILTLMCAATRFPEAFPLRSLRASVIVKEII--KFCSIFGLPKVIQTDQGSNFTSKLFAQV 1059
Query 220 CHRFNIKRCMSSSYH 234
+ MSS+YH
Sbjct 1060 LKELGVSHQMSSAYH 1074
> dre:100331273 RETRotransposon-like family member (retr-1)-like
Length=857
Score = 75.5 bits (184), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 50/185 (27%), Positives = 88/185 (47%), Gaps = 11/185 (5%)
Query 54 GLWRICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCT 113
+++I VP + L+ HDH AGH G KT+ + H++WPG++ Y +C
Sbjct 208 AVYQIVVPT--PYRQHVLYLAHDHQFAGHLGITKTYDRILRHFFWPGLKKDVVKYCRACH 265
Query 114 HCRASKSLNQK-PAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMAD 172
C+ NQ P L + + S H+ +D + LP T +G+ +L ++ ++ +
Sbjct 266 TCQVVGKPNQTIPPAPLIPITVVSEPFQHILVDCVGPLPKTRSGNQFLLTIMCVATRFPE 325
Query 173 FVRAKKSFTAADTVELLADRLIRYHS---FPQVLISDRDPRFQSDLWNQLCHRFNIKRCM 229
+ +K T ++ LI++ S PQV+ +D+ F S L+ Q I +
Sbjct 326 AIPLRKI-----TAPVIIKALIKFFSTFGLPQVIQTDQGTNFLSKLFEQTLQSLEITHRV 380
Query 230 SSSYH 234
SS+YH
Sbjct 381 SSAYH 385
> dre:100330857 LReO_3-like
Length=881
Score = 73.6 bits (179), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 50/182 (27%), Positives = 89/182 (48%), Gaps = 11/182 (6%)
Query 57 RICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCR 116
+I VP E+ L HDH +GH G KT+ + +++WPG++ AY +C+ C+
Sbjct 19 QIVVPV--EYRASILSLAHDHAMSGHLGVTKTYNRVLHNFFWPGLKREVAAYCRTCSVCQ 76
Query 117 ASKSLNQ--KPAGLLEQLLIPSRRR--AHVSLDFITDLPLTTTGHDSILVMVESLSKMAD 172
NQ PA L+ IP+ HV +D + LP T +G++ +L ++ ++ +
Sbjct 77 VMGKPNQVIPPAPLVP---IPAIGEPFEHVIVDCVGPLPKTKSGNEFLLTVMCVATRYPE 133
Query 173 FVRAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSS 232
+ +K T+ V+ L + P+++ +D+ F S L+ Q I+ +SS
Sbjct 134 AIPLRK-ITSKAVVKALV-KFFSTFGLPKIIQTDQGTNFMSKLFTQTFSSLGIQHRVSSP 191
Query 233 YH 234
YH
Sbjct 192 YH 193
> dre:566260 LReO_3-like
Length=1496
Score = 73.6 bits (179), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 50/201 (24%), Positives = 93/201 (46%), Gaps = 6/201 (2%)
Query 34 FCNRQFTFRYLQPYLHIRVHGLWRICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALS 93
CN + L++R + + R+ VP L LH H AGH GQ+KT+A +S
Sbjct 601 LCNEVYVVE--NGVLYVRTNDVLRLVVPSCCRPLV--LHLAHTVPWAGHLGQQKTYARIS 656
Query 94 EHYYWPGMRAYTTAYVESCTHCRASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLT 153
+YWP + + ++C C+ + +++Q+ L+ L + S +++D + L +
Sbjct 657 SRFYWPTLYTDVQTHCKTCAVCQKTSAVSQRGRAPLQPLPVISAPFRRIAMDIVGPLEKS 716
Query 154 TTGHDSILVMVESLSKMADFVRAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQS 213
+ GH ILV+ + ++ + +S T + L R P+ +++D+ F S
Sbjct 717 SAGHRYILVVSDYATRYPE-AYPLRSITTPKIIHALIQLFSRV-GIPEEILTDQGTNFTS 774
Query 214 DLWNQLCHRFNIKRCMSSSYH 234
L QL + I ++ YH
Sbjct 775 RLMGQLHKQMGITAIRTTPYH 795
> dre:100330583 RETRotransposon-like family member (retr-1)-like
Length=1225
Score = 73.6 bits (179), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 45/161 (27%), Positives = 81/161 (50%), Gaps = 3/161 (1%)
Query 75 HDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQK-PAGLLEQLL 133
HDH +GH G KKT+ + +H++WP +++ + +C C+ S NQ P L +
Sbjct 451 HDHDLSGHLGIKKTYQRILKHFFWPRLKSDVAKFCRTCKACQFSGKPNQVIPRAPLVPIP 510
Query 134 IPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFTAADTVELLADRL 193
+ +HV +D + LP T G+ +L ++ + ++ + + +K TA V+ L +
Sbjct 511 VIGEPFSHVIVDCVGPLPKTKAGNQYLLTIMCTATRFPEAIPLRK-ITAPVVVKALV-KF 568
Query 194 IRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
P+V+ +D+ F S L+ Q+ NI SS+YH
Sbjct 569 FSTFGLPRVVQTDQGTNFLSKLFAQVLKTLNISHRTSSAYH 609
> dre:100333332 LReO_3-like
Length=963
Score = 73.2 bits (178), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 45/178 (25%), Positives = 82/178 (46%), Gaps = 8/178 (4%)
Query 59 CVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRAS 118
C PQ + + + H AGH G T + + ++WPG+ Y ++C C+
Sbjct 276 CWPQV-RIIDERRGEKNTHPMAGHLGAANTVKRIRDRFHWPGLDGEVKRYCQACDICQRM 334
Query 119 KSLNQKPAGLLEQLLI--PSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRA 176
P+ L+ +I P R + +D + LP + GH+ ILV+++ ++ + +
Sbjct 335 SPQRPPPSPLITLPIIDVPFTR---IGMDLVGPLPKSARGHEHILVILDYATRYPEAIPL 391
Query 177 KKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
+K+ ++A EL L P +++D+ F S L LCH +K+ +S YH
Sbjct 392 RKATSSAIAKELFL--LCSRVGIPAEILTDQGTPFMSRLMADLCHLLKVKQIKTSVYH 447
> dre:100149641 similar to guanylate binding protein 1, interferon-inducible,
67kDa
Length=1737
Score = 72.8 bits (177), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 58/195 (29%), Positives = 93/195 (47%), Gaps = 17/195 (8%)
Query 47 YLHIRV-HGLWRICVPQFPEFLT----QTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGM 101
Y H+++ G I PE L Q LH H GHQG+++T+ + YWPGM
Sbjct 1291 YRHLKLPGGGEEIDQLLLPEVLQAEVFQQLHGSH-----GHQGRERTYELIRNRCYWPGM 1345
Query 102 RAYTTAYVESCTHCRASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSIL 161
A + C+ C SK LNQ A L+ SR +++DF T L + G + +L
Sbjct 1346 EADVRKRCQECSQCAVSK-LNQPLARAPMGHLLASRPNQILAVDFTT-LERASDGREHVL 1403
Query 162 VMVESLSKMADFV--RAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQL 219
V+ + SK V R +K+ T A+ L+ + R+ P + SD+ F+ + +QL
Sbjct 1404 VITDVFSKYTQAVPTRDQKAITVANI--LIHEWFYRF-GVPAQIHSDQGRNFEGAVVSQL 1460
Query 220 CHRFNIKRCMSSSYH 234
C + +++ + YH
Sbjct 1461 CQLYGVQKTRTVPYH 1475
> dre:561478 LReO_3-like
Length=1553
Score = 72.8 bits (177), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 50/182 (27%), Positives = 89/182 (48%), Gaps = 11/182 (6%)
Query 57 RICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCR 116
+I VP E+ L HDH +GH G KT+ + +++WPG++ AY +C+ C+
Sbjct 691 QIVVPV--EYRASILSLAHDHAMSGHLGVTKTYNRVLHNFFWPGLKREVAAYCRTCSVCQ 748
Query 117 ASKSLNQ--KPAGLLEQLLIPSRRR--AHVSLDFITDLPLTTTGHDSILVMVESLSKMAD 172
NQ PA L+ IP+ HV +D + LP T +G++ +L ++ ++ +
Sbjct 749 VMGKPNQVIPPAPLVP---IPAIGEPFEHVIVDCVGPLPKTKSGNEFLLTVMCVATRYPE 805
Query 173 FVRAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSS 232
+ +K T+ V+ L + P+++ +D+ F S L+ Q I+ +SS
Sbjct 806 AIPLRK-ITSKAVVKALV-KFFSTFGLPKIIQTDQGTNFMSKLFTQTFSSLGIQHRVSSP 863
Query 233 YH 234
YH
Sbjct 864 YH 865
> dre:553442 ccdc33; coiled-coil domain containing 33
Length=1768
Score = 72.4 bits (176), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 52/172 (30%), Positives = 89/172 (51%), Gaps = 13/172 (7%)
Query 65 EFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQK 124
E +TQ LH H GHQG ++T + + YWPGM + + C C+ +K +
Sbjct 869 EVMTQ-LHQQH-----GHQGVERTTQLVWQRCYWPGMSGDIARWCQECERCQCAKGVQPI 922
Query 125 PAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSK--MADFVRAKKSFTA 182
P + +LL +R V+LDF T L + G +++LV+ + SK +A R +++ T
Sbjct 923 PVSFMGRLLA-ARPNEIVALDF-TVLEPSHPGIENVLVITDIFSKYTLAVPTRDQRAETV 980
Query 183 ADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
A L+A+ ++ P + SD+ F+S L QLCH + +++ ++ YH
Sbjct 981 AQV--LVAEWFCKF-GVPGRIHSDQGRNFESVLIQQLCHLYGVEKSRTTPYH 1029
> dre:100329804 zinc finger protein-like
Length=1505
Score = 72.0 bits (175), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 47/173 (27%), Positives = 86/173 (49%), Gaps = 9/173 (5%)
Query 66 FLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQ-- 123
+ + L HDHV AGH G KTF+ ++++++WP +R+ Y SC C+ + NQ
Sbjct 639 YRSAVLKLAHDHVMAGHFGVNKTFSRITKYFFWPSLRSSVGNYCRSCHACQVAGKPNQVV 698
Query 124 KPAGL--LEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFT 181
+PA L + + P R + LD + LP + G+ IL ++ + ++ + V + T
Sbjct 699 RPAPLHPIPVMGEPFER---LILDCVGPLPKSKQGYQYILTLMCAATRFPEAVPLRNIKT 755
Query 182 AADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
+ EL+ + P+V+ +DR F S + + ++ +SS+YH
Sbjct 756 QSIVKELI--KFCSLFGLPRVIQTDRGTNFMSTQFKRALEAISVSHQVSSAYH 806
> dre:100331687 retrovirus polyprotein, putative-like
Length=1341
Score = 72.0 bits (175), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 52/173 (30%), Positives = 85/173 (49%), Gaps = 7/173 (4%)
Query 63 FPEFLTQ-TLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSL 121
PE L L+S HD + GH G ++T + +YWP M +++C+ C KSL
Sbjct 760 LPENLRSFALNSLHDQM--GHMGTERTLDLVRSRFYWPRMGTEVEHKIKTCSRCVRRKSL 817
Query 122 NQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFT 181
QK A L+ + +R V +DF++ P + D ILV+ + +K A +
Sbjct 818 PQKSAPLVN--IQATRPLILVCMDFLSLEPDKSNTRD-ILVITDFFTKYAVAIPTPNQ-K 873
Query 182 AADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
A + L D+ I ++ FP+ L SD+ P F+S +LC I++ ++ YH
Sbjct 874 ARTVAKCLWDQFIVHYGFPERLHSDQGPDFESHTIKELCAIAGIQKIRTTPYH 926
> dre:100148674 Gap-Pol polyprotein-like
Length=1664
Score = 71.6 bits (174), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 53/172 (30%), Positives = 87/172 (50%), Gaps = 13/172 (7%)
Query 65 EFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQK 124
E LTQ LH H GHQG ++T + + YWPGM A + + C C+ +K
Sbjct 1095 EVLTQ-LHQQH-----GHQGVERTSQLVRQRCYWPGMFADIARWCQECERCQCAKGTPSA 1148
Query 125 PAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSK--MADFVRAKKSFTA 182
P+ + LL SR ++LDF P + +G +++LVM + +K +A R +++ T
Sbjct 1149 PSSYMGHLLA-SRPNEILALDFTLMDP-SRSGLENVLVMTDIFTKYTLAIPTRDQRAETV 1206
Query 183 ADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
A L+A+ ++ P + SD+ F+S L QLC + + + +S YH
Sbjct 1207 AQV--LVAEWFCKF-GVPGRIHSDQGRNFESTLIQQLCGLYGVVKSRTSPYH 1255
> dre:100334840 RETRotransposon-like family member (retr-1)-like
Length=1097
Score = 70.9 bits (172), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 47/184 (25%), Positives = 88/184 (47%), Gaps = 5/184 (2%)
Query 52 VHGLWRICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVES 111
+ ++I VP + L HDH +GH G +KT+ + +H++WPG+++ T Y S
Sbjct 304 LDAFYQIVVPA--PYRPHVLCLAHDHPLSGHLGVRKTYNRILKHFFWPGLKSDVTRYCRS 361
Query 112 CTHCRASKSLNQ-KPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKM 170
C C+ + NQ P L+ + + HV +D + + +G+ +L ++ ++
Sbjct 362 CHVCQIAGKPNQVVPPAPLKSIPVLGDPFEHVLIDCVGPFKKSKSGNQFLLTVMCLATRF 421
Query 171 ADFVRAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMS 230
+ + +K TA V+ L + P+++ SD+ F S L+ Q+ I S
Sbjct 422 PEAIPLRK-VTAPVVVKALV-KFFSTFGLPKIVQSDQGTNFMSKLFKQVMQNLGITHRTS 479
Query 231 SSYH 234
S+YH
Sbjct 480 SAYH 483
> dre:100333285 hypothetical protein LOC100333285
Length=340
Score = 70.5 bits (171), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 37/127 (29%), Positives = 70/127 (55%), Gaps = 2/127 (1%)
Query 108 YVESCTHCRASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESL 167
YV++C C+ +K N+KPAG L+Q+ SR +D + +P + ++ +LV V+
Sbjct 8 YVKNCAKCQVTKWDNRKPAGKLQQVTT-SRPNEMWGVDIMGPMPKSGKQNEYLLVFVDYF 66
Query 168 SKMADFVRAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKR 227
SK + + + TA +L ++ + P ++SDR +F S L+ +LC ++NI
Sbjct 67 SKWVELFPMRHA-TAQTIATILRQEMLTWWGVPDFILSDRGAQFVSSLFTELCGKWNITP 125
Query 228 CMSSSYH 234
++++YH
Sbjct 126 KLTTAYH 132
> dre:100004317 RETRotransposon-like family member (retr-1)-like
Length=968
Score = 70.5 bits (171), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 43/161 (26%), Positives = 80/161 (49%), Gaps = 3/161 (1%)
Query 75 HDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQ-KPAGLLEQLL 133
HDH +GH G +KT+ + +H++WPG+++ T Y SC C+ + NQ P L+ +
Sbjct 326 HDHPLSGHLGVRKTYNRILKHFFWPGLKSDVTRYCRSCHVCQIAGKPNQVVPPAPLKPIP 385
Query 134 IPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFTAADTVELLADRL 193
+ HV +D + + +G+ +L ++ ++ + + +K TA V+ L +
Sbjct 386 VLGDPFEHVLIDCVGPFKKSKSGNQFLLTVMCLATRFPEAIPLRK-VTAPVVVKALV-KF 443
Query 194 IRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
P+++ SD+ F S L+ Q+ I SS+YH
Sbjct 444 FSTFGLPKIVQSDQGTNFMSKLFKQVMENLGITHRTSSAYH 484
> dre:100332274 RETRotransposon-like family member (retr-1)-like
Length=949
Score = 70.1 bits (170), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 47/184 (25%), Positives = 88/184 (47%), Gaps = 5/184 (2%)
Query 52 VHGLWRICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVES 111
+ ++I VP + L HDH +GH G +KT+ + +H++WPG+++ T Y S
Sbjct 235 LDAFYQIVVPA--PYRPHVLCLAHDHPLSGHLGVRKTYNRILKHFFWPGLKSDVTRYCRS 292
Query 112 CTHCRASKSLNQ-KPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKM 170
C C+ + NQ P L+ + + HV +D + + +G+ +L ++ ++
Sbjct 293 CHVCQIAGKPNQVVPPAPLKPIPVLGDPFEHVLIDCVGPFKKSKSGNQFLLTVMCLATRF 352
Query 171 ADFVRAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMS 230
+ + +K TA V+ L + P+++ SD+ F S L+ Q+ I S
Sbjct 353 PEAIPLRK-VTAPVVVKALV-KCFSTFGLPKIVQSDQGTNFMSKLFKQVMQNLGITHRTS 410
Query 231 SSYH 234
S+YH
Sbjct 411 SAYH 414
> dre:567586 LReO_3-like
Length=1349
Score = 69.7 bits (169), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 47/168 (27%), Positives = 77/168 (45%), Gaps = 11/168 (6%)
Query 71 LHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCR--ASKSLNQKPAGL 128
L H H AGH G T + + ++WPG+ Y ++C C+ A + P
Sbjct 497 LELAHTHPMAGHLGAANTIQRIRDRFHWPGLNGEVKRYCQACPTCQKTAPQRPPPSPLIP 556
Query 129 LEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFV--RAKKSFTAADTV 186
L + +P R + LD I LP + GH+ ILV+++ ++ + + R S A +
Sbjct 557 LPIIEVPFDR---IGLDLIGPLPKSARGHEHILVILDYATRYPEAIPLRKATSNVIAKEL 613
Query 187 ELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
LL R+ P +++D+ F S L LCH +K+ +S YH
Sbjct 614 FLLCSRV----GIPSEILTDQGTPFMSRLMADLCHLLKVKQLRTSVYH 657
> dre:100151502 retrovirus polyprotein, putative-like; K04228
arginine vasopressin receptor 2
Length=2164
Score = 68.9 bits (167), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 46/171 (26%), Positives = 87/171 (50%), Gaps = 10/171 (5%)
Query 66 FLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQKP 125
+ + L S HD +GH G ++T L + +YWP M +Y YV++C C K++ +K
Sbjct 1138 YWSHVLRSLHDD--SGHLGVERTTELLRDRFYWPRMSSYVEQYVKNCGRCVTRKTVPKKA 1195
Query 126 AGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMAD--FVRAKKSFTAA 183
A L L + V +DF++ P + G ++LV+ + ++ A + +K+ T A
Sbjct 1196 APL--NHLTSNGPFDLVCIDFLSIEP-DSRGLSNVLVVTDHFTRYAQAFVTKDQKALTVA 1252
Query 184 DTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
++L D+ ++ P + SD+ F+S L ++ + I++ +S YH
Sbjct 1253 ---KVLCDKFFVHYGLPTRIHSDQGRDFESGLIKEMLNMLGIRKSRTSPYH 1300
> dre:100334670 retrovirus polyprotein, putative-like
Length=2061
Score = 68.9 bits (167), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 46/171 (26%), Positives = 87/171 (50%), Gaps = 10/171 (5%)
Query 66 FLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQKP 125
+ + L S HD +GH G ++T L + +YWP M +Y YV++C C K++ +K
Sbjct 1441 YWSHVLRSLHDD--SGHLGVERTTELLRDRFYWPRMSSYVEQYVKNCGRCVTRKTVPKKA 1498
Query 126 AGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMAD--FVRAKKSFTAA 183
A L L + V +DF++ P + G ++LV+ + ++ A + +K+ T A
Sbjct 1499 APL--NHLTSNGPFDLVCIDFLSIEP-DSRGLSNVLVVTDHFTRYAQAFVTKDQKALTVA 1555
Query 184 DTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
++L D+ ++ P + SD+ F+S L ++ + I++ +S YH
Sbjct 1556 ---KVLCDKFFVHYGLPTRIHSDQGRDFESGLIKEMLNMLGIRKSRTSPYH 1603
> dre:100000817 LReO_3-like
Length=931
Score = 68.6 bits (166), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 48/180 (26%), Positives = 85/180 (47%), Gaps = 8/180 (4%)
Query 57 RICVP-QFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHC 115
+I VP F E + Q H AGH G +KT+ + +YWPG++ ++ SC C
Sbjct 135 QIVVPLAFRESVMQLAHQG----LAGHTGVRKTYDRIMRQFYWPGVKRDVARFIRSCHTC 190
Query 116 RASKSLNQK-PAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFV 174
+ + NQK P L+ + + S H+ +D + LP + GH +L ++ L+
Sbjct 191 QLTGKPNQKVPTAPLQPIPVTSNPFDHLIIDCVGPLPRSRAGHHYLLTIM-CLTTRYPAA 249
Query 175 RAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
+S T ++ L + + P+V+ SD+ F S + + + +K +SS+YH
Sbjct 250 YPLRSITTKSILKALTN-FMSIFGIPKVIQSDQGSNFMSKQFAKALRQLRVKHNISSAYH 308
> dre:100333139 retrovirus polyprotein, putative-like
Length=2006
Score = 68.6 bits (166), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 46/171 (26%), Positives = 87/171 (50%), Gaps = 10/171 (5%)
Query 66 FLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQKP 125
+ + L S HD +GH G ++T L + +YWP M +Y YV++C C K++ +K
Sbjct 1441 YWSHVLRSLHDD--SGHLGVERTTELLRDRFYWPRMSSYVEQYVKNCGRCVTRKTVPKKA 1498
Query 126 AGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMAD--FVRAKKSFTAA 183
A L L + V +DF++ P + G ++LV+ + ++ A + +K+ T A
Sbjct 1499 APL--NHLTSNGPFDLVCIDFLSIEP-DSRGLSNVLVVTDHFTRYAQAFVTKDQKALTVA 1555
Query 184 DTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
++L D+ ++ P + SD+ F+S L ++ + I++ +S YH
Sbjct 1556 ---KVLCDKFFVHYGLPTRIHSDQGRDFESGLIKEMLNMLGIRKSRTSPYH 1603
Lambda K H
0.325 0.135 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8017665500
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40