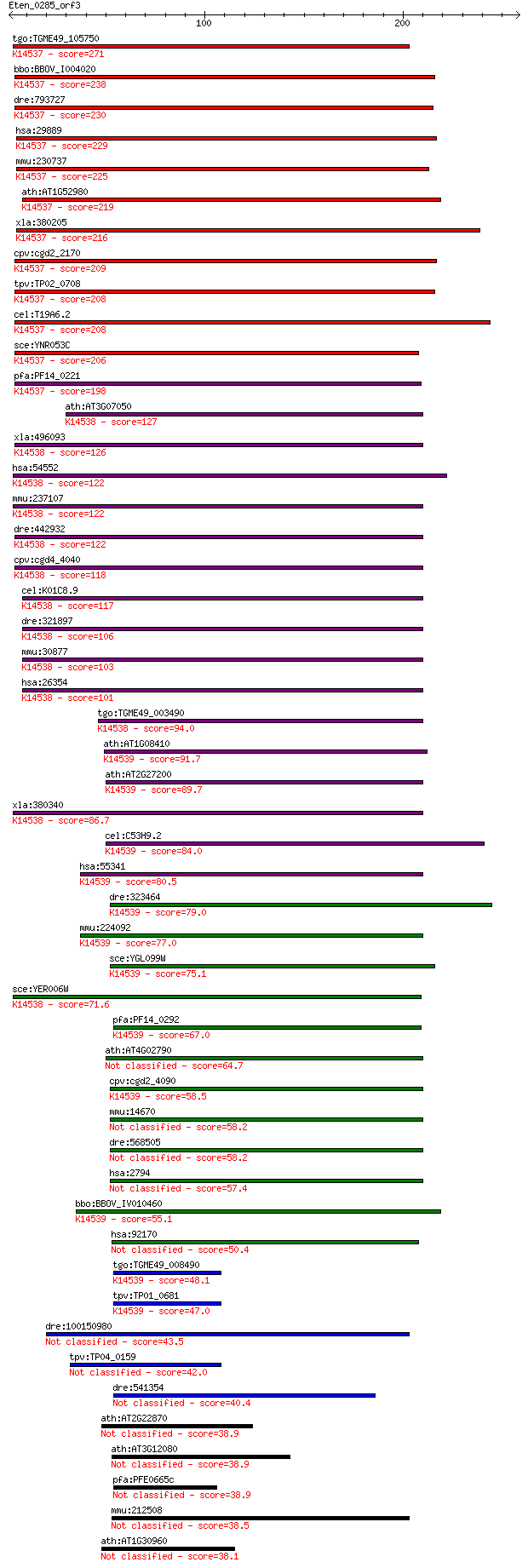
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0285_orf3
Length=258
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_105750 nucleolar GTP-binding protein NOG2, putative... 271 3e-72
bbo:BBOV_I004020 19.m02341; nucleolar GTP-binding protein 2; K... 238 1e-62
dre:793727 gnl2, MGC174441, MGC65800, MGC77319, wu:fc15c12, zg... 230 4e-60
hsa:29889 GNL2, FLJ40906, HUMAUANTIG, NGP1, Ngp-1; guanine nuc... 229 7e-60
mmu:230737 Gnl2, BC003262, HUMAUANTIG, MGC7863, Ngp-1; guanine... 225 1e-58
ath:AT1G52980 GTP-binding family protein; K14537 nuclear GTP-b... 219 1e-56
xla:380205 gnl2, 1i973, MGC52791; guanine nucleotide binding p... 216 5e-56
cpv:cgd2_2170 Ynr053p-like, Yjeq GTpase ; K14537 nuclear GTP-b... 209 7e-54
tpv:TP02_0708 hypothetical protein; K14537 nuclear GTP-binding... 208 1e-53
cel:T19A6.2 ngp-1; Nuclear/nucleolar GTP-binding Protein famil... 208 2e-53
sce:YNR053C NOG2, NUG2; Nog2p; K14537 nuclear GTP-binding protein 206 6e-53
pfa:PF14_0221 GTPase, putative; K14537 nuclear GTP-binding pro... 198 2e-50
ath:AT3G07050 GTP-binding family protein; K14538 nuclear GTP-b... 127 4e-29
xla:496093 gnl3l; guanine nucleotide binding protein-like 3 (n... 126 6e-29
hsa:54552 GNL3L, FLJ10613; guanine nucleotide binding protein-... 122 9e-28
mmu:237107 Gnl3l, BC020354; guanine nucleotide binding protein... 122 1e-27
dre:442932 gnl3l, MGC110536, SI:zK13A21.8, flj10613, flj10613l... 122 1e-27
cpv:cgd4_4040 Yer006wp-like. Yjeq GTpase ; K14538 nuclear GTP-... 118 2e-26
cel:K01C8.9 nst-1; mammalian NucleoSTemin (stem cell marker) r... 117 3e-26
dre:321897 gnl3, MGC123093, id:ibd2914, nstm, wu:fb38a04, wu:f... 106 9e-23
mmu:30877 Gnl3, BC037996, C77032, MGC46970, Ns; guanine nucleo... 103 8e-22
hsa:26354 GNL3, C77032, E2IG3, MGC800, NS; guanine nucleotide ... 101 3e-21
tgo:TGME49_003490 GTPase domain containing protein ; K14538 nu... 94.0 5e-19
ath:AT1G08410 GTP-binding family protein; K14539 large subunit... 91.7 2e-18
ath:AT2G27200 GTP-binding family protein; K14539 large subunit... 89.7 9e-18
xla:380340 gnl3, MGC53851, e2ig3, wu:fc55d07; guanine nucleoti... 86.7 7e-17
cel:C53H9.2 hypothetical protein; K14539 large subunit GTPase ... 84.0 4e-16
hsa:55341 LSG1, FLJ11301, FLJ27294; large subunit GTPase 1 hom... 80.5 5e-15
dre:323464 fb99b06, lsg1, wu:fb99b06; zgc:76988 (EC:3.6.1.-); ... 79.0 2e-14
mmu:224092 Lsg1, 5830465I20, AA409273, D16Bwg1547e; large subu... 77.0 6e-14
sce:YGL099W LSG1, KRE35; Lsg1p; K14539 large subunit GTPase 1 ... 75.1 3e-13
sce:YER006W NUG1; GTPase that associates with nuclear 60S pre-... 71.6 2e-12
pfa:PF14_0292 cytosolic preribosomal GTPase, putative; K14539 ... 67.0 7e-11
ath:AT4G02790 GTP-binding family protein 64.7 3e-10
cpv:cgd2_4090 YawG/Kre35p-like, Yjeq GTpase ; K14539 large sub... 58.5 2e-08
mmu:14670 Gnl1, Gna-rs1, Gnal1; guanine nucleotide binding pro... 58.2 3e-08
dre:568505 gnl1, zgc:154008; guanine nucleotide binding protei... 58.2 3e-08
hsa:2794 GNL1, DKFZp547E038, DKFZp547E128, FLJ37752, HSR1; gua... 57.4 6e-08
bbo:BBOV_IV010460 23.m06487; GTPase subfamily protein; K14539 ... 55.1 2e-07
hsa:92170 MTG1, GTP, GTPBP7, RP11-108K14.2; mitochondrial GTPa... 50.4 6e-06
tgo:TGME49_008490 hypothetical protein ; K14539 large subunit ... 48.1 3e-05
tpv:TP01_0681 hypothetical protein; K14539 large subunit GTPas... 47.0 6e-05
dre:100150980 gtpbpl, MGC109944, MGC192255, MGC86745, zgc:1099... 43.5 8e-04
tpv:TP04_0159 hypothetical protein 42.0 0.002
dre:541354 zgc:113625 40.4 0.007
ath:AT2G22870 EMB2001 (embryo defective 2001); GTP binding 38.9 0.016
ath:AT3G12080 emb2738 (embryo defective 2738); GTP binding 38.9 0.017
pfa:PFE0665c GTP binding protein, putative 38.9 0.021
mmu:212508 Mtg1, Gm169, Gtpbp7, MGC28365; mitochondrial GTPase... 38.5 0.022
ath:AT1G30960 GTP-binding protein (ERG) 38.1 0.035
> tgo:TGME49_105750 nucleolar GTP-binding protein NOG2, putative
; K14537 nuclear GTP-binding protein
Length=641
Score = 271 bits (692), Expect = 3e-72, Method: Compositional matrix adjust.
Identities = 138/200 (69%), Positives = 165/200 (82%), Gaps = 1/200 (0%)
Query 3 QVARYWLQQLSKEMPTLLLQADKNKKNFGRTQLFQLLRQYGQLLSDRKHVSIGFFGYPNV 62
QVAR W+++ SKE+PTL QA K +K GR QLFQLLRQY QL+SDRKHVS+GF GYPNV
Sbjct 294 QVARIWVRRFSKELPTLPFQAKKQEKAAGRLQLFQLLRQYVQLMSDRKHVSVGFIGYPNV 353
Query 63 GKSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLYLIDCPGIVPISSDAGTDTDKVMR 122
GKSSIIN L+SKQVC+AAPI G+TRVWQYVALT +LYLIDCPGIVP S+ +D+ +V+R
Sbjct 354 GKSSIINALRSKQVCRAAPIPGETRVWQYVALTKRLYLIDCPGIVPASASI-SDSLRVIR 412
Query 123 GVVRPERISAPEEHIGTVLEKVKREAIVARYGLDSKISWEDAEEFLSILAVRLGKLKKGG 182
GVVRPERI+ PEEHI VL++V+ E+I ARY L W D+E FL++LA + GKL KGG
Sbjct 413 GVVRPERIACPEEHIDEVLQRVQEESIRARYSLPDACKWTDSESFLAVLAKKKGKLLKGG 472
Query 183 EPDISTAARIMLYDLQRGKL 202
EPDISTAARI+LYDLQRG+L
Sbjct 473 EPDISTAARIVLYDLQRGRL 492
> bbo:BBOV_I004020 19.m02341; nucleolar GTP-binding protein 2;
K14537 nuclear GTP-binding protein
Length=671
Score = 238 bits (607), Expect = 1e-62, Method: Compositional matrix adjust.
Identities = 116/213 (54%), Positives = 153/213 (71%), Gaps = 8/213 (3%)
Query 4 VARYWLQQLSKEMPTLLLQADKNKKNFGRTQLFQLLRQYGQLLSDRKHVSIGFFGYPNVG 63
V W++ L++ +PT+ A K FG+ L QLL+QY QL+ DRKH S+GF GYPNVG
Sbjct 262 VTAAWIKHLNRTIPTVAFHASVTKP-FGKNTLMQLLKQYSQLMKDRKHFSVGFIGYPNVG 320
Query 64 KSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLYLIDCPGIVPI-SSDAGTDTDKVMR 122
KSS+IN LK ++ CKAAPI G+TRVWQYV+LT +++LIDCPG+ PI SD G D++++
Sbjct 321 KSSVINTLKGEKNCKAAPIPGETRVWQYVSLTKRIHLIDCPGVTPIEDSDEG---DRLLK 377
Query 123 GVVRPERISAPEEHIGTVLEKVKREAIVARYGLDSKISWEDAEEFLSILAVRLGKLKKGG 182
GVVR ERIS PE +I VLE + REA+V RYG+ + + FL ++A + GK +KG
Sbjct 378 GVVRVERISDPENYIDRVLEIISREALVKRYGIRDDFT---GDNFLDLVAEKFGKFQKGR 434
Query 183 EPDISTAARIMLYDLQRGKLPYYVLPPCLEAEV 215
PDISTAARI+LYD QRG+LPYY PP +A+V
Sbjct 435 VPDISTAARIVLYDFQRGRLPYYSEPPPSDAKV 467
> dre:793727 gnl2, MGC174441, MGC65800, MGC77319, wu:fc15c12,
zgc:65800, zgc:77319; guanine nucleotide binding protein-like
2 (nucleolar); K14537 nuclear GTP-binding protein
Length=727
Score = 230 bits (586), Expect = 4e-60, Method: Compositional matrix adjust.
Identities = 109/211 (51%), Positives = 151/211 (71%), Gaps = 6/211 (2%)
Query 4 VARYWLQQLSKEMPTLLLQADKNKKNFGRTQLFQLLRQYGQLLSDRKHVSIGFFGYPNVG 63
V ++W+ LS+E PTL A +FG+ L QLLRQ+G+L SD+K +S+GF GYPNVG
Sbjct 264 VTKHWVAVLSQEYPTLAFHASLTN-SFGKGSLIQLLRQFGKLHSDKKQISVGFIGYPNVG 322
Query 64 KSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLYLIDCPGIVPISSDAGTDTDKVMRG 123
KSSIIN L+SK+VC AP+ G+T+VWQY+ L +++LIDCPG+V S D ++TD V++G
Sbjct 323 KSSIINTLRSKKVCNVAPLAGETKVWQYITLMRRIFLIDCPGVVYPSDD--SETDIVLKG 380
Query 124 VVRPERISAPEEHIGTVLEKVKREAIVARYGLDSKISWEDAEEFLSILAVRLGKLKKGGE 183
VV+ E+I PE+HIG VLE+ K E I Y + SW AE+FL LA R GKL KGGE
Sbjct 381 VVQVEKIRNPEDHIGAVLERAKAEYIQKTYRIP---SWSSAEDFLEKLAFRTGKLLKGGE 437
Query 184 PDISTAARIMLYDLQRGKLPYYVLPPCLEAE 214
PD+ T ++++L D QRG++P++V PP +E +
Sbjct 438 PDLPTVSKMVLNDWQRGRIPFFVKPPGVETD 468
> hsa:29889 GNL2, FLJ40906, HUMAUANTIG, NGP1, Ngp-1; guanine nucleotide
binding protein-like 2 (nucleolar); K14537 nuclear
GTP-binding protein
Length=731
Score = 229 bits (584), Expect = 7e-60, Method: Compositional matrix adjust.
Identities = 109/212 (51%), Positives = 150/212 (70%), Gaps = 6/212 (2%)
Query 5 ARYWLQQLSKEMPTLLLQADKNKKNFGRTQLFQLLRQYGQLLSDRKHVSIGFFGYPNVGK 64
+ W+ LS++ PTL A FG+ QLLRQ+G+L +D+K +S+GF GYPNVGK
Sbjct 265 TKRWVAVLSQDYPTLAFHASLTNP-FGKGAFIQLLRQFGKLHTDKKQISVGFIGYPNVGK 323
Query 65 SSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLYLIDCPGIVPISSDAGTDTDKVMRGV 124
SS+IN L+SK+VC API G+T+VWQY+ L +++LIDCPG+V S D+ +TD V++GV
Sbjct 324 SSVINTLRSKKVCNVAPIAGETKVWQYITLMRRIFLIDCPGVVYPSEDS--ETDIVLKGV 381
Query 125 VRPERISAPEEHIGTVLEKVKREAIVARYGLDSKISWEDAEEFLSILAVRLGKLKKGGEP 184
V+ E+I +PE+HIG VLE+ K E I Y +D SWE+AE+FL LA R GKL KGGEP
Sbjct 382 VQVEKIKSPEDHIGAVLERAKPEYISKTYKID---SWENAEDFLEKLAFRTGKLLKGGEP 438
Query 185 DISTAARIMLYDLQRGKLPYYVLPPCLEAEVA 216
D+ T +++L D QRG++P++V PP E VA
Sbjct 439 DLQTVGKMVLNDWQRGRIPFFVKPPNAEPLVA 470
> mmu:230737 Gnl2, BC003262, HUMAUANTIG, MGC7863, Ngp-1; guanine
nucleotide binding protein-like 2 (nucleolar); K14537 nuclear
GTP-binding protein
Length=728
Score = 225 bits (574), Expect = 1e-58, Method: Compositional matrix adjust.
Identities = 106/208 (50%), Positives = 150/208 (72%), Gaps = 6/208 (2%)
Query 5 ARYWLQQLSKEMPTLLLQADKNKKNFGRTQLFQLLRQYGQLLSDRKHVSIGFFGYPNVGK 64
+ W+ LS++ PTL A FG+ QLLRQ+G+L +D+K +S+GF GYPNVGK
Sbjct 265 TKRWVAVLSQDYPTLAFHASLTNP-FGKGAFIQLLRQFGKLHTDKKQISVGFIGYPNVGK 323
Query 65 SSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLYLIDCPGIVPISSDAGTDTDKVMRGV 124
SS+IN L+SK+VC API G+T+VWQY+ L +++LIDCPG+V S D ++TD V++GV
Sbjct 324 SSVINTLRSKKVCNVAPIAGETKVWQYITLMRRIFLIDCPGVVYPSED--SETDIVLKGV 381
Query 125 VRPERISAPEEHIGTVLEKVKREAIVARYGLDSKISWEDAEEFLSILAVRLGKLKKGGEP 184
V+ E+I AP++HIG VLE+ K E I Y ++ SWE+AE+FL LA+R GKL KGGEP
Sbjct 382 VQVEKIKAPQDHIGAVLERAKPEYISKTYKIE---SWENAEDFLEKLALRTGKLLKGGEP 438
Query 185 DISTAARIMLYDLQRGKLPYYVLPPCLE 212
D+ T ++++L D QRG++P++V PP E
Sbjct 439 DMLTVSKMVLNDWQRGRIPFFVKPPNAE 466
> ath:AT1G52980 GTP-binding family protein; K14537 nuclear GTP-binding
protein
Length=576
Score = 219 bits (557), Expect = 1e-56, Method: Compositional matrix adjust.
Identities = 106/211 (50%), Positives = 147/211 (69%), Gaps = 6/211 (2%)
Query 8 WLQQLSKEMPTLLLQADKNKKNFGRTQLFQLLRQYGQLLSDRKHVSIGFFGYPNVGKSSI 67
WL+ LSKE PTL A NK +FG+ L +LRQ+ +L SD++ +S+GF GYPNVGKSS+
Sbjct 267 WLRVLSKEYPTLAFHASVNK-SFGKGSLLSVLRQFARLKSDKQAISVGFVGYPNVGKSSV 325
Query 68 INFLKSKQVCKAAPIRGQTRVWQYVALTSKLYLIDCPGIVPISSDAGTDTDKVMRGVVRP 127
IN L++K VCK API G+T+VWQY+ LT +++LIDCPG+V S D T+TD V++GVVR
Sbjct 326 INTLRTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGVVYQSRD--TETDIVLKGVVRV 383
Query 128 ERISAPEEHIGTVLEKVKREAIVARYGLDSKISWEDAEEFLSILAVRLGKLKKGGEPDIS 187
+ EHIG VL +VK+E + Y + WED +FL L GKL KGGEPD+
Sbjct 384 TNLEDASEHIGEVLRRVKKEHLQRAYKIK---DWEDDHDFLLQLCKSSGKLLKGGEPDLM 440
Query 188 TAARIMLYDLQRGKLPYYVLPPCLEAEVAEA 218
T A+++L+D QRG++P++V PP L+ +E+
Sbjct 441 TGAKMILHDWQRGRIPFFVPPPKLDNVASES 471
> xla:380205 gnl2, 1i973, MGC52791; guanine nucleotide binding
protein-like 2 (nucleolar); K14537 nuclear GTP-binding protein
Length=707
Score = 216 bits (551), Expect = 5e-56, Method: Compositional matrix adjust.
Identities = 108/234 (46%), Positives = 155/234 (66%), Gaps = 8/234 (3%)
Query 5 ARYWLQQLSKEMPTLLLQADKNKKNFGRTQLFQLLRQYGQLLSDRKHVSIGFFGYPNVGK 64
+ W+ LS++ PTL + +FG+ QLLRQ+G+L +D+K +S+GF GYPNVGK
Sbjct 265 TKRWVAILSQDYPTLAFHSSLTN-SFGKGAFIQLLRQFGKLHTDKKQISVGFIGYPNVGK 323
Query 65 SSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLYLIDCPGIVPISSDAGTDTDKVMRGV 124
SS+IN L+SK+VC API G+T+VWQY+ L +++LIDCPG+V S D+ TD V++GV
Sbjct 324 SSVINTLRSKKVCNVAPIAGETKVWQYITLMRRIFLIDCPGVVYPSGDSETDI--VLKGV 381
Query 125 VRPERISAPEEHIGTVLEKVKREAIVARYGLDSKISWEDAEEFLSILAVRLGKLKKGGEP 184
V+ E+I +PE+HI VL++ K E I Y ++ SWE+ E+FL LA R GKL KGGEP
Sbjct 382 VQVEKIKSPEDHIAAVLDRAKPEYIRKTYRIE---SWENPEDFLEKLAFRTGKLLKGGEP 438
Query 185 DISTAARIMLYDLQRGKLPYYVLPPCLEAEVAEAAAEADGTPLDGAMAVPEGDA 238
D T ++++L D QRG++P++V PP A +E A T D ++ DA
Sbjct 439 DRQTVSKMVLNDWQRGRIPFFVKPP--NAPNSEPLQTAQVTTADTSVNKERLDA 490
> cpv:cgd2_2170 Ynr053p-like, Yjeq GTpase ; K14537 nuclear GTP-binding
protein
Length=562
Score = 209 bits (533), Expect = 7e-54, Method: Compositional matrix adjust.
Identities = 100/213 (46%), Positives = 142/213 (66%), Gaps = 3/213 (1%)
Query 4 VARYWLQQLSKEMPTLLLQADKNKKNFGRTQLFQLLRQYGQLLSDRKHVSIGFFGYPNVG 63
VA W+ PT+ + +FG+ LF +LRQY LLSD+KHVS+GF GYPNVG
Sbjct 291 VATKWISFYGSIRPTIAFHSSITN-SFGKRTLFHVLRQYASLLSDKKHVSVGFIGYPNVG 349
Query 64 KSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLYLIDCPGIVPISSDAGTDTDKVMRG 123
KSSIIN L+ +VC API G+T++WQY+ LT ++YLIDCPGIVP + + + V+RG
Sbjct 350 KSSIINTLRGSKVCSVAPIAGETKIWQYIHLTHRIYLIDCPGIVP--PENASSYNVVLRG 407
Query 124 VVRPERISAPEEHIGTVLEKVKREAIVARYGLDSKISWEDAEEFLSILAVRLGKLKKGGE 183
VRPE++S P +I +L VK I +Y L S +W++++EFL+++ RLGK+ +GGE
Sbjct 408 AVRPEKLSDPCIYIKQLLNIVKERHIKEKYNLKSTDNWKNSDEFLTLVGKRLGKVLRGGE 467
Query 184 PDISTAARIMLYDLQRGKLPYYVLPPCLEAEVA 216
D+ T A+I+L D GK+PY++ PP +E A
Sbjct 468 IDLITTAKIILNDWIVGKIPYFIPPPQSSSEFA 500
> tpv:TP02_0708 hypothetical protein; K14537 nuclear GTP-binding
protein
Length=556
Score = 208 bits (530), Expect = 1e-53, Method: Compositional matrix adjust.
Identities = 112/218 (51%), Positives = 147/218 (67%), Gaps = 13/218 (5%)
Query 4 VARYWLQQLSKEMPTLLLQADKNKKNFGRTQLFQLLRQYGQLLSDRKHVSIGFFGYPNVG 63
V W++ L++E+ T+ A K FG+ L QLL+QY QL DRKH S+GF GYPNVG
Sbjct 261 VTAAWIKHLNREITTVAFHASV-KNPFGKNTLIQLLKQYSQLFKDRKHFSVGFIGYPNVG 319
Query 64 KSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLYLIDCPGIVPISSDAGTDTDKVMRG 123
KSS+IN LK + CK API G+TRVWQYV LT +++LIDCPG+ P + G DTDKV++G
Sbjct 320 KSSVINTLKGNRSCKTAPIPGETRVWQYVCLTKRIHLIDCPGVTPF--EEGDDTDKVLKG 377
Query 124 VVRPERISAPEEHIGTVLEKVKREAIVARYGLDSKISWEDAEEFLSILAVRLGKLKKGGE 183
+R ERI PE +I V+E VK++ +V RYG+ K ED FL ++A + GK KKG
Sbjct 378 AIRVERIPDPENYINKVIELVKKDGLVRRYGI--KDFTED--NFLELVAAKFGKFKKGKV 433
Query 184 PDISTAARI------MLYDLQRGKLPYYVLPPCLEAEV 215
PDIS AARI +LYD QRG+LP+Y PP E+++
Sbjct 434 PDISIAARIDEILFLVLYDFQRGRLPFYCEPPKEESKI 471
> cel:T19A6.2 ngp-1; Nuclear/nucleolar GTP-binding Protein family
member (ngp-1); K14537 nuclear GTP-binding protein
Length=651
Score = 208 bits (530), Expect = 2e-53, Method: Compositional matrix adjust.
Identities = 103/240 (42%), Positives = 156/240 (65%), Gaps = 10/240 (4%)
Query 4 VARYWLQQLSKEMPTLLLQADKNKKNFGRTQLFQLLRQYGQLLSDRKHVSIGFFGYPNVG 63
V R W+ +LSKEMPT+ A N +FG+ + LLRQ+ +L DR +S+GF GYPNVG
Sbjct 279 VTRKWIGELSKEMPTIAFHASINN-SFGKGAVINLLRQFAKLHPDRPQISVGFIGYPNVG 337
Query 64 KSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLYLIDCPGIVPISSDAGTDTDKVMRG 123
KSS++N L+ K+VCK API G+T+VWQYV L ++YLID PG+V D ++T +++G
Sbjct 338 KSSLVNTLRKKKVCKTAPIAGETKVWQYVMLMRRIYLIDSPGVVYPQGD--SETQIILKG 395
Query 124 VVRPERISAPEEHIGTVLEKVKREAIVARYGLDSKISWEDAEEFLSILAVRLGKLKKGGE 183
VVR E + PE H+ VL++ K E + +YG+ + D ++FL+ +A++ G+L KGG+
Sbjct 396 VVRVENVKDPENHVQGVLDRCKPEHLRRQYGIP---EFTDVDDFLTKIAIKQGRLLKGGD 452
Query 184 PDISTAARIMLYDLQRGKLPYYVLPPCLEAEVAEAAAEADGTPLDGAMAVPEGDALKLDG 243
PDI ++++L + QRGKLPY+V PP E + ++A P++ M + + L LD
Sbjct 453 PDIVAVSKVVLNEFQRGKLPYFVPPPGCEERAKKDFSQA---PIN-EMCADDDEQLPLDA 508
> sce:YNR053C NOG2, NUG2; Nog2p; K14537 nuclear GTP-binding protein
Length=486
Score = 206 bits (524), Expect = 6e-53, Method: Compositional matrix adjust.
Identities = 101/204 (49%), Positives = 141/204 (69%), Gaps = 5/204 (2%)
Query 4 VARYWLQQLSKEMPTLLLQADKNKKNFGRTQLFQLLRQYGQLLSDRKHVSIGFFGYPNVG 63
VA W++ LSKE PTL A +FG+ L QLLRQ+ QL +DRK +S+GF GYPN G
Sbjct 269 VAAAWVKHLSKERPTLAFHASITN-SFGKGSLIQLLRQFSQLHTDRKQISVGFIGYPNTG 327
Query 64 KSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLYLIDCPGIVPISSDAGTDTDKVMRG 123
KSSIIN L+ K+VC+ API G+T+VWQY+ L +++LIDCPGIVP SS ++ D + RG
Sbjct 328 KSSIINTLRKKKVCQVAPIPGETKVWQYITLMKRIFLIDCPGIVPPSS-KDSEEDILFRG 386
Query 124 VVRPERISAPEEHIGTVLEKVKREAIVARYGLDSKISWEDAEEFLSILAVRLGKLKKGGE 183
VVR E ++ PE++I VL++ + + + Y + W+DA EF+ ILA + G+L KGGE
Sbjct 387 VVRVEHVTHPEQYIPGVLKRCQVKHLERTYEIS---GWKDATEFIEILARKQGRLLKGGE 443
Query 184 PDISTAARIMLYDLQRGKLPYYVL 207
PD S ++ +L D RGK+P++VL
Sbjct 444 PDESGVSKQILNDFNRGKIPWFVL 467
> pfa:PF14_0221 GTPase, putative; K14537 nuclear GTP-binding protein
Length=487
Score = 198 bits (503), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 94/207 (45%), Positives = 142/207 (68%), Gaps = 6/207 (2%)
Query 4 VARYWLQQLSKEMPTLLLQADKNKKNFGRTQLFQLLRQYGQLLSD--RKHVSIGFFGYPN 61
VA W++ LSK+ PT+ A+ NK FG++ LF ++RQY +KH+ IG GYPN
Sbjct 262 VAEKWIKILSKDYPTIAYHANINKP-FGKSDLFNIIRQYTDFFKSQKKKHIHIGLIGYPN 320
Query 62 VGKSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLYLIDCPGIVPISSDAGTDTDKVM 121
VGKS+IIN LK K VC +A I GQT+ WQ++ LT+K+YLIDCPGIVP + D++K++
Sbjct 321 VGKSAIINSLKKKVVCISACIPGQTKYWQFIKLTNKIYLIDCPGIVPYDIE---DSEKIL 377
Query 122 RGVVRPERISAPEEHIGTVLEKVKREAIVARYGLDSKISWEDAEEFLSILAVRLGKLKKG 181
R +R E+I+ P +I + + V I+ Y L ++++++EEFL ILA ++GKL KG
Sbjct 378 RCTMRLEKITNPHFYIDDIFKMVNLSYILKIYKLPEDLTFKNSEEFLEILAKKMGKLLKG 437
Query 182 GEPDISTAARIMLYDLQRGKLPYYVLP 208
GEPDI++ +++++ D +GK+PY+V P
Sbjct 438 GEPDITSVSKVIINDWIKGKIPYFVNP 464
> ath:AT3G07050 GTP-binding family protein; K14538 nuclear GTP-binding
protein
Length=582
Score = 127 bits (319), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 69/180 (38%), Positives = 100/180 (55%), Gaps = 4/180 (2%)
Query 30 FGRTQLFQLLRQYGQLLSDRKHVSIGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVW 89
G L +LL+ Y + +K +++G G PNVGKSS+IN LK V G TR
Sbjct 232 LGADTLIKLLKNYSRSHELKKSITVGIIGLPNVGKSSLINSLKRAHVVNVGATPGLTRSL 291
Query 90 QYVALTSKLYLIDCPGIVPISSDAGTDTDKVMRGVVRPERISAPEEHIGTVLEKVKREAI 149
Q V L + L+DCPG+V + S +G D +R R E++ P + +L+ ++ +
Sbjct 292 QEVHLDKNVKLLDCPGVVMLKS-SGNDASIALRNCKRIEKLDDPVSPVKEILKLCPKDML 350
Query 150 VARYGLDSKISWEDAEEFLSILAVRLGKLKKGGEPDISTAARIMLYDLQRGKLPYYVLPP 209
V Y + S+E ++FL +A GKLKKGG DI AARI+L+D GK+PYY +PP
Sbjct 351 VTLYKIP---SFEAVDDFLYKVATVRGKLKKGGLVDIDAAARIVLHDWNEGKIPYYTMPP 407
> xla:496093 gnl3l; guanine nucleotide binding protein-like 3
(nucleolar)-like; K14538 nuclear GTP-binding protein
Length=561
Score = 126 bits (317), Expect = 6e-29, Method: Compositional matrix adjust.
Identities = 76/231 (32%), Positives = 113/231 (48%), Gaps = 32/231 (13%)
Query 4 VARYWLQQLSKEMPTLLLQADKNKKN-----------------------FGRTQLFQLLR 40
V WL+ L E PT+ +A ++N G L +LL
Sbjct 173 VVEKWLKYLRNEFPTVAFKASTQQQNKNLQQSRVPVKQASEDLLSTGACIGADSLMKLLG 232
Query 41 QYGQLLSDRKHVSIGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLYL 100
Y + + +S+G G+PNVGKSS+IN LK + C G T+ Q V L + L
Sbjct 233 NYCRNKDIKTSISVGVVGFPNVGKSSLINSLKRARACNVGATPGVTKCLQEVHLDKHIKL 292
Query 101 IDCPGIV--PISSDAGTDTDKVMRGVVRPERISAPEEHIGTVLEKVKREAIVARYGLDSK 158
+DCPGIV +SDA ++R V+ E++ P + +L + ++ I+ Y +
Sbjct 293 LDCPGIVMATTTSDAAV----ILRNCVKIEQLVDPVGPVEAILRRCNKDQIIQHYKVS-- 346
Query 159 ISWEDAEEFLSILAVRLGKLKKGGEPDISTAARIMLYDLQRGKLPYYVLPP 209
++ D EFL++LA R GKLKKGG PD AA+ +L D GK+ Y+ PP
Sbjct 347 -NFRDTLEFLAMLAKRQGKLKKGGTPDHEKAAKSVLTDWVSGKISYFTHPP 396
> hsa:54552 GNL3L, FLJ10613; guanine nucleotide binding protein-like
3 (nucleolar)-like; K14538 nuclear GTP-binding protein
Length=582
Score = 122 bits (307), Expect = 9e-28, Method: Compositional matrix adjust.
Identities = 79/243 (32%), Positives = 118/243 (48%), Gaps = 31/243 (12%)
Query 3 QVARYWLQQLSKEMPTLLLQAD-----------------------KNKKNFGRTQLFQLL 39
+V WL L E+PT+ +A K+K FG L ++L
Sbjct 181 EVVEKWLDYLRNELPTVAFKASTQHQVKNLNRCSVPVDQASESLLKSKACFGAENLMRVL 240
Query 40 RQYGQLLSDRKHVSIGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLY 99
Y +L R H+ +G G PNVGKSS+IN LK + C + G T+ Q V L +
Sbjct 241 GNYCRLGEVRTHIRVGVVGLPNVGKSSLINSLKRSRACSVGAVPGITKFMQEVYLDKFIR 300
Query 100 LIDCPGIVP-ISSDAGTDTDKVMRGVVRPERISAPEEHIGTVLEKVKREAIVARYGLDSK 158
L+D PGIVP +S+ GT ++R V ++++ P + T+L++ E I YG+
Sbjct 301 LLDAPGIVPGPNSEVGT----ILRNCVHVQKLADPVTPVETILQRCNLEEISNYYGVS-- 354
Query 159 ISWEDAEEFLSILAVRLGKLKKGGEPDISTAARIMLYDLQRGKLPYYVLPPCLEAEVAEA 218
++ E FL+ +A RLGK KKGG AA+ +L D GK+ +Y+ PP
Sbjct 355 -GFQTTEHFLTAVAHRLGKKKKGGLYSQEQAAKAVLADWVSGKISFYIPPPATHTLPTHL 413
Query 219 AAE 221
+AE
Sbjct 414 SAE 416
> mmu:237107 Gnl3l, BC020354; guanine nucleotide binding protein-like
3 (nucleolar)-like; K14538 nuclear GTP-binding protein
Length=577
Score = 122 bits (307), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 75/232 (32%), Positives = 117/232 (50%), Gaps = 32/232 (13%)
Query 3 QVARYWLQQLSKEMPTLLLQAD------------------------KNKKNFGRTQLFQL 38
++ WL+ L E+PT+ +A K++ FG L ++
Sbjct 174 EIVEKWLEYLLNELPTVAFKASTQHHQVKNLTRCKVPVDQASESLLKSRACFGAENLMRV 233
Query 39 LRQYGQLLSDRKHVSIGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKL 98
L Y +L R H+ +G G PNVGKSS+IN LK + C + G T+ Q V L +
Sbjct 234 LGNYCRLGEVRGHIRVGVVGLPNVGKSSLINSLKRSRACSVGAVPGVTKFMQEVYLDKFI 293
Query 99 YLIDCPGIVP-ISSDAGTDTDKVMRGVVRPERISAPEEHIGTVLEKVKREAIVARYGLDS 157
L+D PGIVP +S+ GT ++R + ++++ P + T+L++ E I + YG+
Sbjct 294 RLLDAPGIVPGPNSEVGT----ILRNCIHVQKLADPVTPVETILQRCNLEEISSYYGVS- 348
Query 158 KISWEDAEEFLSILAVRLGKLKKGGEPDISTAARIMLYDLQRGKLPYYVLPP 209
++ E FL+ +A RLGK KKGG AA+ +L D GK+ +Y LPP
Sbjct 349 --GFQTTEHFLTAVAHRLGKKKKGGVYSQEQAAKAVLADWVSGKISFYTLPP 398
> dre:442932 gnl3l, MGC110536, SI:zK13A21.8, flj10613, flj10613l,
si:dkey-13a21.8, zgc:110536; guanine nucleotide binding
protein-like 3 (nucleolar)-like; K14538 nuclear GTP-binding
protein
Length=565
Score = 122 bits (306), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 70/229 (30%), Positives = 113/229 (49%), Gaps = 28/229 (12%)
Query 4 VARYWLQQLSKEMPTLLLQADKNKKN-----------------------FGRTQLFQLLR 40
+ W++ L E PT+ ++ ++N G L +LL
Sbjct 173 IVEKWIKYLRNEFPTVAFKSSTQQQNKNLKRSRVPVTQATQELLESSACVGADCLMKLLG 232
Query 41 QYGQLLSDRKHVSIGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLYL 100
Y + + +++G G+PNVGKSS+IN LK + C G T+ Q V L + L
Sbjct 233 NYCRNQDIKTAITVGVVGFPNVGKSSLINSLKRARACNVGATPGVTKCLQEVHLDKHIKL 292
Query 101 IDCPGIVPISSDAGTDTDKVMRGVVRPERISAPEEHIGTVLEKVKREAIVARYGLDSKIS 160
+DCPGIV +S +D ++R V+ E++ P + +L + + I+ YG+
Sbjct 293 LDCPGIVMATS--TSDAAMILRNCVKIEQLVDPLPAVEAILRRCNKMQIIDHYGIP---D 347
Query 161 WEDAEEFLSILAVRLGKLKKGGEPDISTAARIMLYDLQRGKLPYYVLPP 209
++ A EFL++LA R GKL+KGG PD AA+ +L D G++ Y+ PP
Sbjct 348 FQTAHEFLALLARRQGKLRKGGLPDSDKAAKSVLMDWTGGRISYFTHPP 396
> cpv:cgd4_4040 Yer006wp-like. Yjeq GTpase ; K14538 nuclear GTP-binding
protein
Length=478
Score = 118 bits (295), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 76/232 (32%), Positives = 113/232 (48%), Gaps = 31/232 (13%)
Query 4 VARYWLQQLSKEMPTLLLQADKNKKN-------------------------FGRTQLFQL 38
V + WL L +E PTL ++ N FG + L L
Sbjct 213 VVKEWLTYLRREHPTLAFKSALNSSTEFGVNHSKSSGLNASHDFIKASSVAFGVSPLMSL 272
Query 39 LRQYGQLLSD-RKHVSIGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSK 97
++ Y + + +K ++IG GYPNVGKSS+IN LK K + G TR Q + L S
Sbjct 273 IKNYSRYNKNSKKSITIGVMGYPNVGKSSLINSLKRGYCVKVGAVAGVTRHLQRIDLDST 332
Query 98 LYLIDCPGIVPISSDAGTDTDKVMRGVVRPERISAPEEHIGTVLEKVKREAIVARYGLDS 157
LID PG+V + D +V+R V+ + E I +L+K+ E ++ Y +
Sbjct 333 TKLIDSPGVV--FTGNSQDPSQVLRNTVQLTNVKDYFEPISLLLQKIDHEILLKLYKIP- 389
Query 158 KISWEDAEEFLSILAVRLGKLKKGGEPDISTAARIMLYDLQRGKLPYYVLPP 209
+ + EFL+ +++ GKL KGG PDI++AA I+L D GK+ YY PP
Sbjct 390 --IFNNVSEFLTNVSISRGKLNKGGIPDINSAAMIVLTDWFNGKISYYNFPP 439
> cel:K01C8.9 nst-1; mammalian NucleoSTemin (stem cell marker)
related family member (nst-1); K14538 nuclear GTP-binding protein
Length=556
Score = 117 bits (294), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 66/221 (29%), Positives = 110/221 (49%), Gaps = 22/221 (9%)
Query 8 WLQQLSKEMPTLLLQAD--KNKKNFGRTQ-----------------LFQLLRQYGQLLSD 48
WL+ L + PT+ +A + K N GR + ++L Y +
Sbjct 197 WLEYLRGQFPTIAFKASTQEQKSNIGRFNSAILNNTETSKCVGADIVMKILANYCRNKDI 256
Query 49 RKHVSIGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLYLIDCPGIVP 108
+ + +G G+PNVGKSS+IN LK ++ C + G T+ Q V L + LID PG++
Sbjct 257 KTSIRVGVVGFPNVGKSSVINSLKRRKACNVGNLPGITKEIQEVELDKNIRLIDSPGVIL 316
Query 109 ISSDAGTDTDKVMRGVVRPERISAPEEHIGTVLEKVKREAIVARYGLDSKISWEDAEEFL 168
+S + ++ +R + + P + +L + +E I+ Y L + ++FL
Sbjct 317 VSQKDLDPIEVALKNAIRVDNLLDPIAPVHAILRRCSKETIMLHYNL---ADFNSVDQFL 373
Query 169 SILAVRLGKLKKGGEPDISTAARIMLYDLQRGKLPYYVLPP 209
+ LA R+GKL++G PD++ AA+ +L D GKL YY PP
Sbjct 374 AQLARRIGKLRRGARPDVNAAAKRVLNDWNTGKLRYYTHPP 414
> dre:321897 gnl3, MGC123093, id:ibd2914, nstm, wu:fb38a04, wu:fc55d07,
zgc:123093; guanine nucleotide binding protein-like
3 (nucleolar); K14538 nuclear GTP-binding protein
Length=561
Score = 106 bits (264), Expect = 9e-23, Method: Compositional matrix adjust.
Identities = 69/226 (30%), Positives = 104/226 (46%), Gaps = 29/226 (12%)
Query 8 WLQQLSKEMPTLLLQADKNKKN------------------------FGRTQLFQLLRQYG 43
WL L E PT L ++ K+ FG+ L Q L
Sbjct 194 WLHFLEAECPTFLFKSSMQLKDRTVQQKRQQRGTNAVLDHSRAASCFGKDFLLQTLNDLA 253
Query 44 QLLSDRKHVSIGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLYLIDC 103
+ +G G+PNVGKSSIIN LK + C A RG TR Q V +T K+ +ID
Sbjct 254 NKKEGETMLKVGVVGFPNVGKSSIINSLKEMRACNAGVQRGLTRCMQEVHITKKVKMIDS 313
Query 104 PGIVPISSDAGTDTDKVMRGVVRPERISAPEEHIGTVLEKVKREAIVARYGLDSKISWED 163
PGI+ S+ G+ +R + E+ +P+E + +L++ ++ ++ +Y + +
Sbjct 314 PGILAALSNPGSAM--ALRSLQVEEKEESPQEAVRNLLKQCNQQHVMLQYNVP---DYRS 368
Query 164 AEEFLSILAVRLGKLKKGGEPDISTAARIMLYDLQRGKLPYYVLPP 209
+ EFL+ A++ G L+KGG D AA L D KL YY P
Sbjct 369 SLEFLTTFAMKHGLLQKGGVADTELAATTFLNDWTGAKLSYYSRVP 414
> mmu:30877 Gnl3, BC037996, C77032, MGC46970, Ns; guanine nucleotide
binding protein-like 3 (nucleolar); K14538 nuclear GTP-binding
protein
Length=538
Score = 103 bits (256), Expect = 8e-22, Method: Compositional matrix adjust.
Identities = 69/221 (31%), Positives = 108/221 (48%), Gaps = 27/221 (12%)
Query 8 WLQQLSKEMPTLLLQADKNKKN-------------------FGRTQLFQLLRQYGQLLSD 48
WL L+KE+PT++ +A N KN G+ L++LL + Q S
Sbjct 189 WLNYLNKELPTVVFKASTNLKNRKTFKIKKKKVVPFQSKICCGKEALWKLLGDFQQ--SC 246
Query 49 RKHVSIGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLYLIDCPGIVP 108
K + +G G+PNVGKSS+IN LK + +C G TR Q V L ++ +ID P +
Sbjct 247 GKDIQVGVIGFPNVGKSSVINSLKQEWICNVGISMGLTRSMQIVPLDKQITIIDSPCL-- 304
Query 109 ISSDAGTDTDKVMRGVVRPERISAPEEHIGTVLEKVKREAIVARYGLDSKISWEDAEEFL 168
I S + T +R E + P E +L + E +V +Y + ++D+ F
Sbjct 305 IISPCNSPTALALRSPASIEELR-PLEAASAILSQADNEQVVLKYTVP---EYKDSLHFF 360
Query 169 SILAVRLGKLKKGGEPDISTAARIMLYDLQRGKLPYYVLPP 209
+ LA R G +KGG P++ +AA+++ + L YY PP
Sbjct 361 TKLAQRRGLHQKGGSPNVESAAKLVWSEWTGASLGYYCHPP 401
> hsa:26354 GNL3, C77032, E2IG3, MGC800, NS; guanine nucleotide
binding protein-like 3 (nucleolar); K14538 nuclear GTP-binding
protein
Length=549
Score = 101 bits (251), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 70/226 (30%), Positives = 110/226 (48%), Gaps = 34/226 (15%)
Query 8 WLQQLSKEMPTLLLQAD-------------KNKKN---------FGRTQLFQLLRQYGQL 45
WL L KE+PT++ +A K KKN FG+ L++LL + +
Sbjct 191 WLNYLKKELPTVVFRASTKPKDKGKITKRVKAKKNAAPFRSEVCFGKEGLWKLLGGFQET 250
Query 46 LSDRKHVSIGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLYLIDCPG 105
S K + +G G+PNVGKSSIIN LK +Q+C G TR Q V L ++ +ID P
Sbjct 251 CS--KAIRVGVIGFPNVGKSSIINSLKQEQMCNVGVSMGLTRSMQVVPLDKQITIIDSPS 308
Query 106 IV--PISSDAGTDTDKVMRGVVRPERISAPEEHIGTVLEKVKREAIVARYGLDSKISWED 163
+ P++S + +R E + P E +L + +V +Y + + +
Sbjct 309 FIVSPLNSSSAL----ALRSPASIE-VVKPMEAASAILSQADARQVVLKYTVP---GYRN 360
Query 164 AEEFLSILAVRLGKLKKGGEPDISTAARIMLYDLQRGKLPYYVLPP 209
+ EF ++LA R G +KGG P++ AA+++ + L YY PP
Sbjct 361 SLEFFTVLAQRRGMHQKGGIPNVEGAAKLLWSEWTGASLAYYCHPP 406
> tgo:TGME49_003490 GTPase domain containing protein ; K14538
nuclear GTP-binding protein
Length=673
Score = 94.0 bits (232), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 63/195 (32%), Positives = 94/195 (48%), Gaps = 36/195 (18%)
Query 46 LSDRKHVSIGFFGYPNVGKSSIINFLKSKQVCKAAPIR---GQTRVWQYVALTSKLYLID 102
++ + ++ +G GYPNVGKSS++N L C AA + G T+ QYV + + LID
Sbjct 299 VTAKANLLVGVVGYPNVGKSSLVNAL--THSCSAAAVAAMPGSTKTLQYVRIDKHIQLID 356
Query 103 CPGIV---------------PISSDAGTDTDK-------------VMRGVVRPERISAPE 134
PG++ P S+ A D ++R ++ R+ P+
Sbjct 357 SPGVLFSPSKNAQDDAVLLPPPSARASACQDIPRAEAAAESCSAFILRSLLPVHRLENPQ 416
Query 135 EHIGTVLEKVKREAIVARYGLDSKISWEDAEEFLSILAVRLGKLKKGGEPDISTAARIML 194
E V E + Y L+ + +E L++LA R GKLKKGG PD+ AA+I L
Sbjct 417 EVATGVAAWCCSETLQRLYRLE---GFSSPQEMLALLAHRRGKLKKGGIPDLDAAAKIFL 473
Query 195 YDLQRGKLPYYVLPP 209
D Q G +PYY +PP
Sbjct 474 QDWQSGVIPYYTMPP 488
> ath:AT1G08410 GTP-binding family protein; K14539 large subunit
GTPase 1 [EC:3.6.1.-]
Length=589
Score = 91.7 bits (226), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 58/171 (33%), Positives = 88/171 (51%), Gaps = 9/171 (5%)
Query 49 RKHVSIGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLYLIDCPGIVP 108
R +GF GYPNVGKSS IN L ++ G+T+ +Q + ++ +L L DCPG+V
Sbjct 306 RDQAVVGFVGYPNVGKSSTINALVGQKRTGVTSTPGKTKHFQTLIISDELMLCDCPGLV- 364
Query 109 ISSDAGTDTDKVMRGVVRPERISAPEEHIGTVLEKVKREAIVARYG--LDSKISWE---- 162
S + + + + GV+ +R++ E I V +KV R I + Y L ++E
Sbjct 365 FPSFSSSRYEMIASGVLPIDRMTEHREAIQVVADKVPRRVIESVYNISLPKPKTYERQSR 424
Query 163 --DAEEFLSILAVRLGKLKKGGEPDISTAARIMLYDLQRGKLPYYVLPPCL 211
A E L G + G PD + AAR++L D GKLP+Y +PP +
Sbjct 425 PPHAAELLKSYCASRGYVASSGLPDETKAARLILKDYIGGKLPHYAMPPGM 475
> ath:AT2G27200 GTP-binding family protein; K14539 large subunit
GTPase 1 [EC:3.6.1.-]
Length=537
Score = 89.7 bits (221), Expect = 9e-18, Method: Compositional matrix adjust.
Identities = 60/168 (35%), Positives = 86/168 (51%), Gaps = 9/168 (5%)
Query 50 KHVSIGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLYLIDCPGIVPI 109
+ V +GF GYPNVGKSS IN L ++ G+T+ +Q + ++ L L DCPG+V
Sbjct 303 EQVVVGFVGYPNVGKSSTINALVGQKRTGVTSTPGKTKHFQTLIISEDLMLCDCPGLV-F 361
Query 110 SSDAGTDTDKVMRGVVRPERISAPEEHIGTVLEKVKREAI--VARYGLDSKISWED---- 163
S + + + V GV+ +R++ E I V E V R AI V L S+E
Sbjct 362 PSFSSSRYEMVASGVLPIDRMTEHLEAIKVVAELVPRHAIEDVYNISLPKPKSYEPQSRP 421
Query 164 --AEEFLSILAVRLGKLKKGGEPDISTAARIMLYDLQRGKLPYYVLPP 209
A E L + G + G PD + AAR +L D GKLP++ +PP
Sbjct 422 PLASELLRTYCLSRGYVASSGLPDETRAARQILKDYIEGKLPHFAMPP 469
> xla:380340 gnl3, MGC53851, e2ig3, wu:fc55d07; guanine nucleotide
binding protein-like 3 (nucleolar); K14538 nuclear GTP-binding
protein
Length=542
Score = 86.7 bits (213), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 65/235 (27%), Positives = 106/235 (45%), Gaps = 43/235 (18%)
Query 3 QVARYWLQQLSKEMPTLLLQA-----DKNK-----------------KNFGRTQLFQLLR 40
++ WLQ LS E+PT+ + DK++ K G L ++L
Sbjct 179 EMVEKWLQVLSAELPTVPFRCVAQIQDKSEKKKKKKVPVSADLVTDPKCPGGQVLLKIL- 237
Query 41 QYGQLLSDRKHVSIGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLYL 100
+ S + + +G G+ NVGKSS+IN LK VC P +G T+ Q V L ++ L
Sbjct 238 -HSLCPSHNEAIKVGVIGFANVGKSSVINSLKQSHVCNVGPSKGTTKFLQEVRLDPQIRL 296
Query 101 IDCPGIVPISSDAGTDTDKVMRGVVRPERISAPEEHIGT------VLEKVKREAIVARYG 154
+D P + + S ++R SA E + +L+ ++ ++ Y
Sbjct 297 LDSPAL--LVSPHNPPVALMLR--------SASESKVDVLAAVEAILKHCSKQELMLHYT 346
Query 155 LDSKISWEDAEEFLSILAVRLGKLKKGGEPDISTAARIMLYDLQRGKLPYYVLPP 209
+ + ++ E L++LA R G LKKGG PD A R++ D ++ YY PP
Sbjct 347 I---ADYRNSLECLTLLAHRRGMLKKGGVPDTEGAGRLLFNDWMGARMKYYCRPP 398
> cel:C53H9.2 hypothetical protein; K14539 large subunit GTPase
1 [EC:3.6.1.-]
Length=554
Score = 84.0 bits (206), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 65/204 (31%), Positives = 98/204 (48%), Gaps = 17/204 (8%)
Query 50 KHVSIGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLYLIDCPGIVPI 109
K V +G GYPNVGKSS IN L + + G+TR +Q + + S+L L DCPG+V
Sbjct 297 KPVMVGMVGYPNVGKSSTINKLAGGKKVSVSATPGKTRHFQTIHIDSQLCLCDCPGLVMP 356
Query 110 SSDAGTDTDKVMRGVVRPERISAPEEHIGT---VLEKVKREAIVARYGLD-SKISWEDAE 165
S G ++ + G++ +++ +H G +L +V I A Y + ++ A
Sbjct 357 SFSFGR-SEMFLNGILPVDQM---RDHFGPTSLLLSRVPVHVIEATYSIMLPEMQSPSAI 412
Query 166 EFLSILAVRLGKLKKGGEPDISTAARIMLYDLQRGKLPYYVLPPCLEAEVAEAAAEADGT 225
L+ LA G + G PD S AAR+M D+ GKL + PP +E E + + D
Sbjct 413 NLLNSLAFMRGFMASSGIPDCSRAARLMFKDVVSGKLIWAAAPPGVEQEEFDRISYPDKK 472
Query 226 PLD---------GAMAVPEGDALK 240
D + + EGD LK
Sbjct 473 TRDIGRVQMEKLAKLQLLEGDELK 496
> hsa:55341 LSG1, FLJ11301, FLJ27294; large subunit GTPase 1 homolog
(S. cerevisiae); K14539 large subunit GTPase 1 [EC:3.6.1.-]
Length=658
Score = 80.5 bits (197), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 54/185 (29%), Positives = 88/185 (47%), Gaps = 13/185 (7%)
Query 37 QLLRQYGQLLSDRK----HVSIGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVWQYV 92
+LL + +L + RK +++G GYPNVGKSS IN + + + G T+ +Q +
Sbjct 368 ELLELFKELHTGRKVKDGQLTVGLVGYPNVGKSSTINTIMGNKKVSVSATPGHTKHFQTL 427
Query 93 ALTSKLYLIDCPGIVPISSDAGTDTDKVMRGVVRPERISAPEEHIGTVLEKVKREAIVAR 152
+ L L DCPG+V + S T + G++ +++ + V + + R + A
Sbjct 428 YVEPGLCLCDCPGLV-MPSFVSTKAEMTCSGILPIDQMRDHVPPVSLVCQNIPRHVLEAT 486
Query 153 YGLDSKISWED--------AEEFLSILAVRLGKLKKGGEPDISTAARIMLYDLQRGKLPY 204
YG++ ED +EE L+ G + G+PD +AR +L D GKL Y
Sbjct 487 YGINIITPREDEDPHRPPTSEELLTAYGYMRGFMTAHGQPDQPRSARYILKDYVSGKLLY 546
Query 205 YVLPP 209
PP
Sbjct 547 CHPPP 551
> dre:323464 fb99b06, lsg1, wu:fb99b06; zgc:76988 (EC:3.6.1.-);
K14539 large subunit GTPase 1 [EC:3.6.1.-]
Length=640
Score = 79.0 bits (193), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 58/201 (28%), Positives = 90/201 (44%), Gaps = 17/201 (8%)
Query 52 VSIGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLYLIDCPGIVPISS 111
+++G GYPNVGKSS IN + + + G T+ +Q + + L L DCPG+V + S
Sbjct 369 ITVGLVGYPNVGKSSTINTIFRNKKVSVSATPGHTKHFQTLFVEPGLCLCDCPGLV-MPS 427
Query 112 DAGTDTDKVMRGVVRPERISAPEEHIGTVLEKVKREAIVARYGLDSKISWEDA------- 164
T + + G++ +++ I V + + R + YG++ ED
Sbjct 428 FVSTKAEMICSGILPIDQMRDHVPAISLVCQNIPRNVLEGTYGINIIRPREDEDPDRPPT 487
Query 165 -EEFLSILAVRLGKLKKGGEPDISTAARIMLYDLQRGKLPYYVLPPCLEAEVAEAAAEAD 223
EE L G + G+PD S +AR +L D GKL Y PP + E D
Sbjct 488 YEELLMAYGYMRGFMTAHGQPDQSRSARYVLKDYVSGKLLYCHPPPHINPE--------D 539
Query 224 GTPLDGAMAVPEGDALKLDGS 244
P A+ A ++DGS
Sbjct 540 FQPQHAKFAMRITGAEQIDGS 560
> mmu:224092 Lsg1, 5830465I20, AA409273, D16Bwg1547e; large subunit
GTPase 1 homolog (S. cerevisiae); K14539 large subunit
GTPase 1 [EC:3.6.1.-]
Length=644
Score = 77.0 bits (188), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 52/185 (28%), Positives = 88/185 (47%), Gaps = 13/185 (7%)
Query 37 QLLRQYGQLLSDRK----HVSIGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVWQYV 92
+LL + +L + +K +++G GYPNVGKSS IN + + + G T+ +Q +
Sbjct 354 ELLELFKKLHTGKKVKDGQLTVGLVGYPNVGKSSTINTIMGNKKVSVSATPGHTKHFQTL 413
Query 93 ALTSKLYLIDCPGIVPISSDAGTDTDKVMRGVVRPERISAPEEHIGTVLEKVKREAIVAR 152
+ L L DCPG+V + S T + + G++ +++ + V + + R +
Sbjct 414 YVEPGLCLCDCPGLV-MPSFVSTKAEMICNGILPIDQMRDHVPPVSLVCQNIPRRVLEVT 472
Query 153 YGLDSKISWED--------AEEFLSILAVRLGKLKKGGEPDISTAARIMLYDLQRGKLPY 204
YG++ ED +EE L+ G + G+PD +AR +L D GKL Y
Sbjct 473 YGINIIKPREDEDPYRPPTSEELLTAYGCMRGFMTAHGQPDQPRSARYILKDYVGGKLLY 532
Query 205 YVLPP 209
PP
Sbjct 533 CHPPP 537
> sce:YGL099W LSG1, KRE35; Lsg1p; K14539 large subunit GTPase
1 [EC:3.6.1.-]
Length=640
Score = 75.1 bits (183), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 56/174 (32%), Positives = 84/174 (48%), Gaps = 11/174 (6%)
Query 52 VSIGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLYLIDCPGIVPISS 111
++IG GYPNVGKSS IN L + + G+T+ +Q + L+ + L DCPG+V +
Sbjct 337 INIGLVGYPNVGKSSTINSLVGAKKVSVSSTPGKTKHFQTIKLSDSVMLCDCPGLV-FPN 395
Query 112 DAGTDTDKVMRGVVRPERISAPEEHIGTVLEKVKREAIVARYGLDSKISWED-------- 163
A + V GV+ +++ G V E++ + I A YG+ + D
Sbjct 396 FAYNKGELVCNGVLPIDQLRDYIGPAGLVAERIPKYYIEAIYGIHIQTKSRDEGGNGDIP 455
Query 164 -AEEFLSILAVRLGKLKKG-GEPDISTAARIMLYDLQRGKLPYYVLPPCLEAEV 215
A+E L A G + +G G D A+R +L D GKL Y PP LE +
Sbjct 456 TAQELLVAYARARGYMTQGYGSADEPRASRYILKDYVNGKLLYVNPPPHLEDDT 509
> sce:YER006W NUG1; GTPase that associates with nuclear 60S pre-ribosomes,
required for export of 60S ribosomal subunits from
the nucleus; K14538 nuclear GTP-binding protein
Length=520
Score = 71.6 bits (174), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 71/238 (29%), Positives = 110/238 (46%), Gaps = 35/238 (14%)
Query 3 QVARYWLQQLSKEMPTLLLQADK---NKKNFGR--------TQLFQLLRQYGQLLSDRKH 51
V WL L PT+ L+A N +F R + L + L+ Y + ++
Sbjct 221 HVLEQWLNYLKSSFPTIPLRASSGAVNGTSFNRKLSQTTTASALLESLKTYSNNSNLKRS 280
Query 52 VSIGFFGYPNVGKSSIINFLKSKQ--VCKAAPI---RGQTRVWQYVALTSKLYLIDCPGI 106
+ +G GYPNVGKSS+IN L +++ KA P+ G T + + + +KL ++D PGI
Sbjct 281 IVVGVIGYPNVGKSSVINALLARRGGQSKACPVGNEAGVTTSLREIKIDNKLKILDSPGI 340
Query 107 VPISSDAGTDTDKVMRGVVRPERISAPEEHI----GTVLEKVKR--------EAIVARYG 154
S + KV + P +HI VL VKR E+ Y
Sbjct 341 CFPSEN--KKRSKVEHEAELALLNALPAKHIVDPYPAVLMLVKRLAKSDEMTESFKKLYE 398
Query 155 LDSKISWEDAE----EFLSILAVRLGKLKKGGEPDISTAARIMLYDLQRGKLPYYVLP 208
+ I DA+ FL +A + G+L KGG P++++A +L D + GK+ +VLP
Sbjct 399 I-PPIPANDADTFTKHFLIHVARKRGRLGKGGIPNLASAGLSVLNDWRDGKILGWVLP 455
> pfa:PF14_0292 cytosolic preribosomal GTPase, putative; K14539
large subunit GTPase 1 [EC:3.6.1.-]
Length=833
Score = 67.0 bits (162), Expect = 7e-11, Method: Composition-based stats.
Identities = 51/170 (30%), Positives = 77/170 (45%), Gaps = 16/170 (9%)
Query 54 IGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVWQYVALTS-KLYLIDCPGIVPISSD 112
IGF G+PNVGKSSIIN L K+ + G+T+ +Q + L L DCPG++ S
Sbjct 601 IGFIGFPNVGKSSIINCLIGKKKVSVSRQPGKTKHFQTITLKHFPFSLCDCPGLI-FPSL 659
Query 113 AGTDTDKVMRGVVRPERISAPEEHIGTVLEKVKREAIVARYGLDSKISWE---------- 162
D ++ GV + + ++ + + Y +D+ I +
Sbjct 660 VFNKNDLIINGVFSIDHFKGDVVTLVQIICNIIPFKLCNNYKIDTNIIHQYLNEKGHISY 719
Query 163 --DAEEFLSILAV--RLGKLKKGGEPDISTAARIMLYDLQRGKLPYYVLP 208
DA EFL + KGG+ + S A RI+++D GKL Y LP
Sbjct 720 FLDASEFLKKFCTFRKFVSGGKGGQLNFSHATRIIIHDFISGKLLYNFLP 769
> ath:AT4G02790 GTP-binding family protein
Length=372
Score = 64.7 bits (156), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 53/168 (31%), Positives = 83/168 (49%), Gaps = 16/168 (9%)
Query 50 KHVSIGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLYLIDCPGIVPI 109
+ V G GYPNVGKSS+IN L +++C AAP G TR ++V L L L+D PG++P+
Sbjct 212 RSVRAGIIGYPNVGKSSLINRLLKRKICAAAPRPGVTREMKWVKLGKDLDLLDSPGMLPM 271
Query 110 SSDAGTDTDKVMRGVVRPERISAPEEHIGTVLEKVKR------EAIVARYGLDSKISWED 163
D K+ E+ + G +++ + R +A+ RY + ++
Sbjct 272 RIDDQAAAIKLAICDDIGEKAYDFTDVAGILVQMLARIPEVGAKALYNRYKI--QLEGNC 329
Query 164 AEEFLSILAVRLGKLKKGGEPDISTAARIMLYDLQRGKLPYYVL--PP 209
++F+ L + L GG D AA +L D ++GK Y L PP
Sbjct 330 GKKFVKTLGLNLF----GG--DSHQAAFRILTDFRKGKFGYVSLERPP 371
> cpv:cgd2_4090 YawG/Kre35p-like, Yjeq GTpase ; K14539 large subunit
GTPase 1 [EC:3.6.1.-]
Length=666
Score = 58.5 bits (140), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 56/181 (30%), Positives = 84/181 (46%), Gaps = 31/181 (17%)
Query 52 VSIGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSK-----------LYL 100
++IG G+PNVGKSSI+N L Q + G+T+ Q + L + L
Sbjct 417 LTIGMVGFPNVGKSSIVNALFGSQKSSISRTPGKTKHLQTLRLKPPHLNDKEEDQDFITL 476
Query 101 IDCPGIVPISSDAGTDTDKVMRGVVRPERISAPEEHI-GTVLEKVKR--EAIVARY---- 153
DCPG+V + S T ++ GV P +H G L+ ++ E I A+
Sbjct 477 CDCPGLV-MPSFTSTKEHLLINGV-------TPIDHFRGNFLDTIQLIGERITAQLYKTY 528
Query 154 --GLDSKI-SWEDAEEFLSILAV--RLGKLKKGGEPDISTAARIMLYDLQRGKLPYYVLP 208
G+D ++ ++ +FL+ L L + KG PD S A R++L D GKL Y P
Sbjct 529 FDGIDYQVPRIFNSTQFLNKLCETRHLFQQGKGAIPDWSKAGRMILRDYWSGKLLYCHTP 588
Query 209 P 209
P
Sbjct 589 P 589
> mmu:14670 Gnl1, Gna-rs1, Gnal1; guanine nucleotide binding protein-like
1
Length=607
Score = 58.2 bits (139), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 49/163 (30%), Positives = 75/163 (46%), Gaps = 7/163 (4%)
Query 52 VSIGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLYLIDCPGIVPISS 111
V+IG G+PNVGKSS+IN L ++V + G TR +Q LT + L DCPG+ I
Sbjct 361 VTIGCIGFPNVGKSSLINGLVGRKVVSVSRTPGHTRYFQTYFLTPSVKLCDCPGL--IFP 418
Query 112 DAGTDTDKVMRGVVRPERISAPEEHIGTVLEKVKREAIVA-RYGLDSKISWED---AEEF 167
+V+ G+ +I P +G + ++ +A++ R+ S E A +
Sbjct 419 SLLPRQLQVLAGIYPIAQIQEPYTSVGYLASRIPVQALLHLRHPEAEDPSAEHPWCAWDI 478
Query 168 LSILAVRLG-KLKKGGEPDISTAARIMLYDLQRGKLPYYVLPP 209
A + G K K D+ AA +L G+L PP
Sbjct 479 CEAWAEKRGYKTAKAARNDVYRAANSLLRLAVDGRLSLCFYPP 521
> dre:568505 gnl1, zgc:154008; guanine nucleotide binding protein-like
1
Length=602
Score = 58.2 bits (139), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 48/169 (28%), Positives = 76/169 (44%), Gaps = 15/169 (8%)
Query 52 VSIGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLYLIDCPGIV-PIS 110
+++G G+PNVGKSS++N L ++V + G T+ +Q LT + L DCPG+V P
Sbjct 349 LTLGCIGFPNVGKSSVLNSLVGRKVVSVSRTPGHTKYFQTYYLTPTVKLCDCPGLVFPSR 408
Query 111 SDAGTDTDKVMRGVVRPERISAPEEHIGTVLEKVKREAIVARYGLD---------SKISW 161
D +V+ G+ ++ P +G + E+ +I+ D S W
Sbjct 409 VDKQL---QVLAGIYPVSQLQEPYSSVGHLCERTNYLSILKLTHPDHSPETPHDPSTQDW 465
Query 162 EDAEEFLSILAVRLG-KLKKGGEPDISTAARIMLYDLQRGKLPYYVLPP 209
A + A R G K K D+ AA +L G+L + PP
Sbjct 466 T-AWDVCEAWAERRGYKTAKAARNDVYRAANSLLRLAIDGRLCLCLRPP 513
> hsa:2794 GNL1, DKFZp547E038, DKFZp547E128, FLJ37752, HSR1; guanine
nucleotide binding protein-like 1
Length=607
Score = 57.4 bits (137), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 49/163 (30%), Positives = 75/163 (46%), Gaps = 7/163 (4%)
Query 52 VSIGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSKLYLIDCPGIVPISS 111
V+IG G+PNVGKSS+IN L ++V + G TR +Q LT + L DCPG+ I
Sbjct 361 VTIGCVGFPNVGKSSLINGLVGRKVVSVSRTPGHTRYFQTYFLTPSVKLCDCPGL--IFP 418
Query 112 DAGTDTDKVMRGVVRPERISAPEEHIGTVLEKVKREAIVA-RYGLDSKISWED---AEEF 167
+V+ G+ +I P +G + ++ +A++ R+ S E A +
Sbjct 419 SLLPRQLQVLAGIYPIAQIQEPYTAVGYLASRIPVQALLHLRHPEAEDPSAEHPWCAWDI 478
Query 168 LSILAVRLG-KLKKGGEPDISTAARIMLYDLQRGKLPYYVLPP 209
A + G K K D+ AA +L G+L PP
Sbjct 479 CEAWAEKRGYKTAKAARNDVYRAANSLLRLAVDGRLSLCFHPP 521
> bbo:BBOV_IV010460 23.m06487; GTPase subfamily protein; K14539
large subunit GTPase 1 [EC:3.6.1.-]
Length=826
Score = 55.1 bits (131), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 51/196 (26%), Positives = 83/196 (42%), Gaps = 15/196 (7%)
Query 35 LFQLLRQYGQLLSDRKHVSIGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVWQYVAL 94
+F LRQ + SD ++G GYPNVGKSS+IN L + G+T+ Q +AL
Sbjct 547 IFPELRQ--EDTSDMPVYTVGCVGYPNVGKSSLINCLMEVTKTNVSCQPGKTKHLQTLAL 604
Query 95 TS-KLYLIDCPGIVPISSDAGTDTDKVMRGVVRPERISAPEEHIGTVLEKVKREAIVARY 153
+ L DCPG++ + ++ +V ++ RY
Sbjct 605 KKYNITLCDCPGLI-FPNIVANKHHLLVNSIVSTAHFRGSLIFAVQLICNRIPNQCCERY 663
Query 154 GLD---------SKISWEDAEEFLSIL--AVRLGKLKKGGEPDISTAARIMLYDLQRGKL 202
+D +K + +FL + + + KGG+PD+ AA++++ D G L
Sbjct 664 DVDRAECITINKNKKPILLSTKFLECICNSRKFFSGGKGGQPDLGRAAKLVVKDYVNGNL 723
Query 203 PYYVLPPCLEAEVAEA 218
Y PP + V E
Sbjct 724 LYCAWPPDFKPSVVEC 739
> hsa:92170 MTG1, GTP, GTPBP7, RP11-108K14.2; mitochondrial GTPase
1 homolog (S. cerevisiae)
Length=334
Score = 50.4 bits (119), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 51/177 (28%), Positives = 80/177 (45%), Gaps = 24/177 (13%)
Query 53 SIGFFGYPNVGKSSIINFLKSKQVCKAAPIR-----GQTR-VWQYVALTSK--LYLIDCP 104
I G PNVGKSS+IN L+ + + K R G TR V + ++ + ++L+D P
Sbjct 145 CIMVIGVPNVGKSSLINSLRRQHLRKGKATRVGGEPGITRAVMSKIQVSERPLMFLLDTP 204
Query 105 GIV-PISSDAGTDTDKVMRGVVRPERI--SAPEEHIGTVLEKVKREAIVARYGLDSKISW 161
G++ P T + G V + +++ L K +R V YGL S +
Sbjct 205 GVLAPRIESVETGLKLALCGTVLDHLVGEETMADYLLYTLNKHQRFGYVQHYGLGS--AC 262
Query 162 EDAEEFLSILAVRLGKLKKGG-----------EPDISTAARIMLYDLQRGKLPYYVL 207
++ E L +AV+LGK +K +P+ AAR L +RG L +L
Sbjct 263 DNVERVLKSVAVKLGKTQKVKVLTGTGNVNIIQPNYPAAARDFLQTFRRGLLGSVML 319
> tgo:TGME49_008490 hypothetical protein ; K14539 large subunit
GTPase 1 [EC:3.6.1.-]
Length=1064
Score = 48.1 bits (113), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 23/55 (41%), Positives = 33/55 (60%), Gaps = 1/55 (1%)
Query 54 IGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVWQYVAL-TSKLYLIDCPGIV 107
+G G+PNVGKSS+IN L + + G+TR Q + + + L L DCPG+V
Sbjct 669 VGLVGFPNVGKSSVINALLGSKKVSVSRTPGKTRHLQTLVVGDTGLTLCDCPGLV 723
> tpv:TP01_0681 hypothetical protein; K14539 large subunit GTPase
1 [EC:3.6.1.-]
Length=529
Score = 47.0 bits (110), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 25/56 (44%), Positives = 35/56 (62%), Gaps = 3/56 (5%)
Query 54 IGFFGYPNVGKSSIIN-FLKSKQVCKAAPIRGQTRVWQYVAL-TSKLYLIDCPGIV 107
+GF GYPNVGKSS+IN ++S + C G+T+ Q + L S + L DCPG +
Sbjct 447 VGFVGYPNVGKSSLINCLMESTRTCVGTQ-PGKTKHIQTLPLKNSDIILCDCPGTI 501
> dre:100150980 gtpbpl, MGC109944, MGC192255, MGC86745, zgc:109944,
zgc:86745; GTP binding protein like
Length=248
Score = 43.5 bits (101), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 54/215 (25%), Positives = 95/215 (44%), Gaps = 34/215 (15%)
Query 20 LLQADKNKKNFG--RTQLFQLLRQYGQLL--SDRKH------VSIGFFGYPNVGKSSIIN 69
L Q D+N K R +FQ++ +L+ + R H + G PNVGKSS+IN
Sbjct 30 LRQHDENVKKHPVIRCLMFQIVPLVSKLIESTSRFHREEERCYCLMVIGVPNVGKSSLIN 89
Query 70 -----FLKSKQVCKAAPIRGQTR-VWQYVALTSK--LYLIDCPGIVPIS-SDAGTDTDKV 120
+LK + K G T+ V + + + ++L+D PG++P + T
Sbjct 90 ALRRTYLKKGKASKVGAEPGITKAVLTKIQVCERPIIHLLDTPGVLPPRIENIETGMKLA 149
Query 121 MRGVVRPERI--SAPEEHIGTVLEKVKREAIVARYGLDSKISWEDAEEFLSILAVRLGKL 178
+ G V + + + L +++R + V +Y L+ +D + L +AV+LGK
Sbjct 150 LCGTVLDHLVGEDVIADFLLFSLNRLERFSYVEKYSLEE--PCDDIQHVLKCIAVKLGKT 207
Query 179 KKGGE-----------PDISTAARIMLYDLQRGKL 202
++ PD S AA + ++G+L
Sbjct 208 RRVKAITGVGNITVQLPDYSAAAYDFIRAFRKGEL 242
> tpv:TP04_0159 hypothetical protein
Length=538
Score = 42.0 bits (97), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 29/89 (32%), Positives = 46/89 (51%), Gaps = 14/89 (15%)
Query 32 RTQLFQLLRQYGQL-----LSDRKHVSIGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQT 86
R +F++ R+ + L+ R+ + + G PNVGKSS++N L ++V K+ + G T
Sbjct 290 RKHIFRMCRRVNERRLRKGLNPRE-IRVILMGMPNVGKSSLVNRLLGRKVTKSYNVPGLT 348
Query 87 RVWQYVALTSK--------LYLIDCPGIV 107
+ T+K L LID PGIV
Sbjct 349 KNINIYKSTTKSNTIKNKRLLLIDTPGIV 377
> dre:541354 zgc:113625
Length=313
Score = 40.4 bits (93), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 36/137 (26%), Positives = 67/137 (48%), Gaps = 10/137 (7%)
Query 54 IGFFGYPNVGKSSIINFLKSKQVCKAAPIRG---QTRVWQYVALTSKLYLIDCPGIVPIS 110
I G VGKS++ N + +++V ++ P Q+ V T ++Y+ID PGI+
Sbjct 67 IVMIGKTGVGKSAVGNTILNREVFESKPSANSITQSCRKASVYDTREIYVIDTPGIL--- 123
Query 111 SDAGTDTDKVMRGVVRPERISAPEEHIGTVLEKVKREAIVARYGLDS--KISWEDAEEFL 168
D + D + R +V+ ++SAP H ++ ++ R + + + ++ EDA ++
Sbjct 124 -DTSKEKDIIKREIVKCIKVSAPGPHAFLLVIQIGRFTAEEQRAVQALQELFGEDASNYM 182
Query 169 SILAVRLGKLKKGGEPD 185
+L G L KG D
Sbjct 183 IVLFTH-GDLLKGQTID 198
> ath:AT2G22870 EMB2001 (embryo defective 2001); GTP binding
Length=300
Score = 38.9 bits (89), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 26/79 (32%), Positives = 41/79 (51%), Gaps = 5/79 (6%)
Query 48 DRKHVSIGFFGYPNVGKSSIINFL-KSKQVCKAAPIRGQTRVWQYVALTSKLYLIDCP-- 104
DR ++I G NVGKSS+IN L + K+V + G+T++ + + Y++D P
Sbjct 119 DRPEIAI--LGRSNVGKSSLINCLVRKKEVALTSKKPGKTQLINHFLVNKSWYIVDLPGY 176
Query 105 GIVPISSDAGTDTDKVMRG 123
G +S A TD +G
Sbjct 177 GFAKVSDAAKTDWSAFTKG 195
> ath:AT3G12080 emb2738 (embryo defective 2738); GTP binding
Length=587
Score = 38.9 bits (89), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 31/94 (32%), Positives = 41/94 (43%), Gaps = 4/94 (4%)
Query 53 SIGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVWQYVALT----SKLYLIDCPGIVP 108
+I G PNVGKSSI+N L + +P+ G TR T K LID GI
Sbjct 370 AIAIIGRPNVGKSSILNALVREDRTIVSPVSGTTRDAIDAEFTGPDGEKFRLIDTAGIRK 429
Query 109 ISSDAGTDTDKVMRGVVRPERISAPEEHIGTVLE 142
SS A + + V R R + + V+E
Sbjct 430 KSSVASSGSTTEAMSVNRAFRAIRRSDVVALVIE 463
> pfa:PFE0665c GTP binding protein, putative
Length=286
Score = 38.9 bits (89), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 22/53 (41%), Positives = 31/53 (58%), Gaps = 1/53 (1%)
Query 54 IGFFGYPNVGKSSIIN-FLKSKQVCKAAPIRGQTRVWQYVALTSKLYLIDCPG 105
I FG NVGKSS+IN L ++V +A+ G+TR L + L ++D PG
Sbjct 79 IAIFGRSNVGKSSLINALLNYREVSQASNTPGRTRHLFIFNLLNYLSIVDLPG 131
> mmu:212508 Mtg1, Gm169, Gtpbp7, MGC28365; mitochondrial GTPase
1 homolog (S. cerevisiae)
Length=326
Score = 38.5 bits (88), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 48/175 (27%), Positives = 78/175 (44%), Gaps = 30/175 (17%)
Query 53 SIGFFGYPNVGKSSIINFLKSKQV--CKAAPIRGQTRVWQYVALTSKL--------YLID 102
I G PNVGKSS+IN L+ + + KAA + G+ + + A+TS++ +L+D
Sbjct 144 CIMVVGVPNVGKSSLINSLRRQHLRTGKAARVGGEPGITR--AVTSRIQVCERPLVFLLD 201
Query 103 CPGIV-PISSDAGTDTDKVMRGVVRPERISAPEEHIGTVLEKVKREAI---VARYGLDSK 158
PG++ P T + G V + + E +L + R + V Y L S
Sbjct 202 TPGVLAPRIESVETGLKLALCGTVL-DHLVGEETMADYLLYTLNRHGLFGYVQHYALAS- 259
Query 159 ISWEDAEEFLSILAVRLGKLKKGG-----------EPDISTAARIMLYDLQRGKL 202
+ + E L +A++L K +K +PD + AAR L + G L
Sbjct 260 -ACDQIEWVLKNVAIKLRKTRKVKVLTGTGNVNVIQPDYAMAARDFLRTFRSGLL 313
> ath:AT1G30960 GTP-binding protein (ERG)
Length=437
Score = 38.1 bits (87), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 37/71 (52%), Gaps = 5/71 (7%)
Query 48 DRKHVSIGFFGYPNVGKSSIINFLKSKQVCKAAPIRGQTRVWQYVALTSK----LYLIDC 103
D+K +++G G PN GKSS+ NF+ +V AA + T + + + +K + D
Sbjct 150 DQKSLNVGIIGPPNAGKSSLTNFMVGTKVA-AASRKTNTTTHEVLGVLTKGDTQVCFFDT 208
Query 104 PGIVPISSDAG 114
PG++ S G
Sbjct 209 PGLMLKKSGYG 219
Lambda K H
0.319 0.136 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9468570544
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40