bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0288_orf3
Length=324
Score E
Sequences producing significant alignments: (Bits) Value
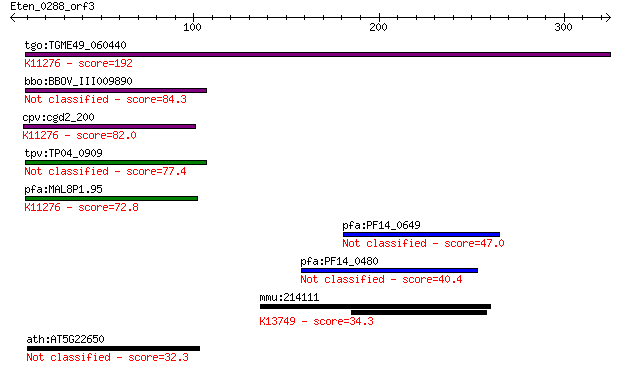
tgo:TGME49_060440 46 kDa FK506-binding nuclear protein, putati... 192 1e-48
bbo:BBOV_III009890 17.m07857; hypothetical protein 84.3 6e-16
cpv:cgd2_200 hypothetical protein ; K11276 nucleophosmin 1 82.0
tpv:TP04_0909 hypothetical protein 77.4 6e-14
pfa:MAL8P1.95 conserved Plasmodium protein, unknown function; ... 72.8 2e-12
pfa:PF14_0649 conserved Plasmodium protein, unknown function 47.0 1e-04
pfa:PF14_0480 conserved Plasmodium protein, unknown function 40.4 0.008
mmu:214111 Slc24a1, MGC27617; solute carrier family 24 (sodium... 34.3 0.69
ath:AT5G22650 HD2B; HD2B (HISTONE DEACETYLASE 2B); histone dea... 32.3 2.8
> tgo:TGME49_060440 46 kDa FK506-binding nuclear protein, putative
(EC:5.2.1.8); K11276 nucleophosmin 1
Length=311
Score = 192 bits (488), Expect = 1e-48, Method: Compositional matrix adjust.
Identities = 144/327 (44%), Positives = 203/327 (62%), Gaps = 27/327 (8%)
Query 9 MMFARNLKPGETVKVSAEDGGEVLHLSQACLLRPADAGRTYLKVVQGKEAFAACVLQKDR 68
M + +KPG+TV +S EDGGEVLHLSQ C+ +P D GRTY++V+Q + ++ C+LQKD+
Sbjct 1 MFYGVVVKPGQTVTLSPEDGGEVLHLSQVCMPQPKDGGRTYVQVIQEGKTYSVCMLQKDK 60
Query 69 LECCSLDLFLSARQGLLLQLEG-KSEVSLLGYFEPEADSAAEDEAGFAAAAAAAAFEPDS 127
LE SLDLFLS RQG+ ++ EG K+EV L GYFEP+A+ DE + +
Sbjct 61 LEFTSLDLFLSTRQGITIKTEGGKNEVHLTGYFEPDAEGLDSDEDEEESESDM------E 114
Query 128 EDDSQEASEDEPGRMAAPVVRDGRVEDLDAEEEDTKKKGKVSSKKGA------AALKALQ 181
E+ A + P + G++EDLD +E+ + + + A +KAL
Sbjct 115 ENVMHHARKHIP-------TKGGKIEDLDGNDEEEEDEDEDEDDDDEDDDEDDAEMKALT 167
Query 182 AAAANDDEDDEDDEDDEDDEDDDEEGSEDESGEEDEEESEEEEEAPKQQQQQ--DKKRKQ 239
A ++D++DE++E +DD++D E G ++S +ED+EE EEEE +P+ ++ DKKRK
Sbjct 168 AGDDDEDDEDENEEGSDDDDEDMENGIGEDSEDEDDEEDEEEEASPQNKKASVGDKKRKA 227
Query 240 QQQQKQQQQPNKKAKNQGGEANTSS-SGDEASYRNALVDFLKKNGPTPLASLGQKVKKPA 298
+ Q NKKAK + + GD A+Y++ALVDFLKKNG TPLA+LGQKVKKPA
Sbjct 228 VEVA---QASNKKAKPSAAPSKGAPQKGDAAAYKSALVDFLKKNGKTPLATLGQKVKKPA 284
Query 299 SLP-KMAAFLKANSDKFAVEGGKCSLK 324
+ KMA FLKAN+ F V G CSLK
Sbjct 285 GVSEKMAQFLKANASTFDVSNGVCSLK 311
> bbo:BBOV_III009890 17.m07857; hypothetical protein
Length=139
Score = 84.3 bits (207), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 40/99 (40%), Positives = 63/99 (63%), Gaps = 2/99 (2%)
Query 9 MMFARNLKPGETVKVSAEDGGEVLHLSQACLLRPADAGRTYLKVVQGKEAFAACVLQKDR 68
M+F + PG +V E ++H+SQ CL P ++ +TY++++ G ++F CVLQKD
Sbjct 1 MLFGAVIAPGSSVTPKNELA-NIVHISQVCLNDPKNSEKTYVQLIDGNKSFNLCVLQKDV 59
Query 69 LECCSLDLFLSARQGLLLQLE-GKSEVSLLGYFEPEADS 106
E S+D+F S G+ L + GK+EV ++GYFEPE +S
Sbjct 60 CEHASIDIFFSTAGGIKLSTKGGKNEVHVVGYFEPEEES 98
> cpv:cgd2_200 hypothetical protein ; K11276 nucleophosmin 1
Length=323
Score = 82.0 bits (201), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 40/93 (43%), Positives = 57/93 (61%), Gaps = 1/93 (1%)
Query 8 KMMFARNLKPGETVKVSAEDGGEVLHLSQACLLRPADAGRTYLKVVQGKEAFAACVLQKD 67
KM +KPG+ VK+ + GE+LHLSQACL P D GR Y++ + A+ C LQK
Sbjct 13 KMFCGLIIKPGQKVKLDSTQ-GEILHLSQACLSEPKDNGRVYVQAIDNGNAYTICSLQKG 71
Query 68 RLECCSLDLFLSARQGLLLQLEGKSEVSLLGYF 100
+E +LDLFLS + L + GK+EV + G++
Sbjct 72 TIEHANLDLFLSTSAEIELSIIGKNEVHISGFY 104
> tpv:TP04_0909 hypothetical protein
Length=138
Score = 77.4 bits (189), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 43/99 (43%), Positives = 57/99 (57%), Gaps = 2/99 (2%)
Query 9 MMFARNLKPGETVKVSAEDGGEVLHLSQACLLRPADAGRTYLKVVQGKEAFAACVLQKDR 68
M+F L PG TV E ++HLSQ CL P RTY+++V G + + C LQKD
Sbjct 1 MLFGAVLAPGATVTPKNELA-NIVHLSQVCLNEPKSDERTYVQLVDGNKVYNLCSLQKDV 59
Query 69 LECCSLDLFLSARQGLLLQLE-GKSEVSLLGYFEPEADS 106
E +LDLF S L L + G +EV ++GYFEPE D+
Sbjct 60 NEHATLDLFFSTTGDLKLTTKGGPNEVHVIGYFEPEDDA 98
> pfa:MAL8P1.95 conserved Plasmodium protein, unknown function;
K11276 nucleophosmin 1
Length=315
Score = 72.8 bits (177), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 36/94 (38%), Positives = 54/94 (57%), Gaps = 1/94 (1%)
Query 9 MMFARNLKPGETVKVSAEDGGEVLHLSQACLLRPADAGRTYLKVVQGKEAFAACVLQKDR 68
M + + +KP ETV +DG V+HLS+ACL P G+ Y++V G + C LQK+
Sbjct 1 MFYGKVVKPDETVVPEVKDGYSVIHLSRACLNNPEHEGKIYVQVEDGNGCYNICCLQKNV 60
Query 69 LECCSLDLFLSARQGLLLQLEGKS-EVSLLGYFE 101
E LD+FL + ++ G S EV ++GY+E
Sbjct 61 CEDTPLDIFLMLDNDVKIKTSGSSNEVHIVGYYE 94
> pfa:PF14_0649 conserved Plasmodium protein, unknown function
Length=2558
Score = 47.0 bits (110), Expect = 1e-04, Method: Composition-based stats.
Identities = 16/84 (19%), Positives = 52/84 (61%), Gaps = 0/84 (0%)
Query 181 QAAAANDDEDDEDDEDDEDDEDDDEEGSEDESGEEDEEESEEEEEAPKQQQQQDKKRKQQ 240
Q ++++D+ DE+ ++++D++++ ++DE+ +E+++ES+ E + Q +++++ +
Sbjct 2037 QNENQDENQDENQDENQDENQDENQDENQDENRDENQDESQNENQNENQDDNENEEKPNE 2096
Query 241 QQQKQQQQPNKKAKNQGGEANTSS 264
+ +++PN + EA+ +
Sbjct 2097 GSNENEEKPNDGSIENNEEADKEA 2120
> pfa:PF14_0480 conserved Plasmodium protein, unknown function
Length=1653
Score = 40.4 bits (93), Expect = 0.008, Method: Composition-based stats.
Identities = 31/96 (32%), Positives = 51/96 (53%), Gaps = 8/96 (8%)
Query 158 EEEDTKKKGKVSSKKGAAALKALQAAAANDDEDDEDDEDDEDDEDDDEEGS-EDESGEED 216
E+EDT K K KG K + +D +D+ EDD ++D E S ED+S E+D
Sbjct 986 EKEDTWKNQK---HKGINKHK--DDSHEDDSHEDDSHEDDSHEDDSHENDSHEDDSHEDD 1040
Query 217 EEESEEEEEAPKQQQQQDKKRKQQQQQKQQQQPNKK 252
E++ EE K+Q +K+ ++ ++K + NK+
Sbjct 1041 SHENDSHEE--KEQYIYNKELYEKIKKKPGKNMNKE 1074
> mmu:214111 Slc24a1, MGC27617; solute carrier family 24 (sodium/potassium/calcium
exchanger), member 1; K13749 solute carrier
family 24 (sodium/potassium/calcium exchanger), member
1
Length=1130
Score = 34.3 bits (77), Expect = 0.69, Method: Composition-based stats.
Identities = 31/139 (22%), Positives = 62/139 (44%), Gaps = 15/139 (10%)
Query 136 EDEPGRMAAPVVRDGRVEDLDAEEED---TKKKGKVSSKKGAAALKALQAAAANDDEDDE 192
EDEP + A V D ++ED ++ + ++G + + + + DE
Sbjct 680 EDEPAELPAVTVTPAPAPDAKGDQEDDPGCQEDVDEAERRGEMTGEEGEKETETEGKKDE 739
Query 193 DDEDDE------------DDEDDDEEGSEDESGEEDEEESEEEEEAPKQQQQQDKKRKQQ 240
+ + E + E ++EG + G+EDE+E E E E K +Q+ + + + +
Sbjct 740 QEGETEAERKEDEQEEETEAEGKEQEGETEAEGKEDEQEGETEAEGKKDEQEGETEAEGK 799
Query 241 QQQKQQQQPNKKAKNQGGE 259
++Q+ + + K Q GE
Sbjct 800 EEQEGETEAEGKEDEQEGE 818
Score = 33.5 bits (75), Expect = 1.00, Method: Composition-based stats.
Identities = 20/85 (23%), Positives = 45/85 (52%), Gaps = 12/85 (14%)
Query 185 ANDDEDDEDDEDDEDDEDDDEEGSED------------ESGEEDEEESEEEEEAPKQQQQ 232
A ED+++ E + + + D++EG + G+EDE+E E E E ++Q+
Sbjct 770 AEGKEDEQEGETEAEGKKDEQEGETEAEGKEEQEGETEAEGKEDEQEGETEAEGKEEQEG 829
Query 233 QDKKRKQQQQQKQQQQPNKKAKNQG 257
+ + ++ +Q+++ + K K++G
Sbjct 830 ETEAESKEVEQERETEAEGKDKHEG 854
> ath:AT5G22650 HD2B; HD2B (HISTONE DEACETYLASE 2B); histone deacetylase
Length=305
Score = 32.3 bits (72), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 24/94 (25%), Positives = 39/94 (41%), Gaps = 3/94 (3%)
Query 10 MFARNLKPGETVKVSAEDGGEVLHLSQACLLRPADAGRTY-LKVVQGKEAFAACVLQKDR 68
+ + P KV+ E+ ++H+SQA L +G + L V G L +D+
Sbjct 3 FWGVAVTPKNATKVTPEED-SLVHISQASLDCTVKSGESVVLSVTVGGAKLVIGTLSQDK 61
Query 69 LECCSLDLFLSARQGLLLQLEGKSEVSLLGYFEP 102
S DL ++ L K+ V +GY P
Sbjct 62 FPQISFDLVFD-KEFELSHSGTKANVHFIGYKSP 94
Lambda K H
0.304 0.124 0.331
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 13394984740
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40