bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0364_orf1
Length=139
Score E
Sequences producing significant alignments: (Bits) Value
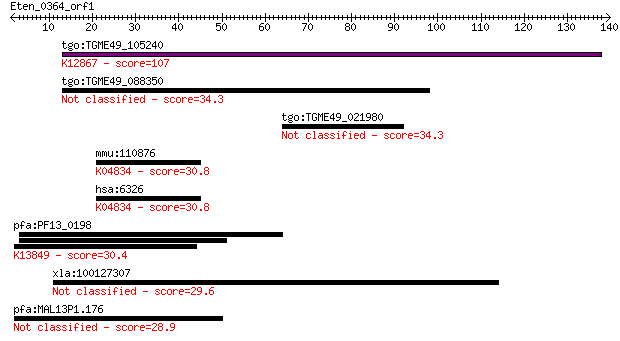
tgo:TGME49_105240 XPA-binding protein, putative ; K12867 pre-m... 107 1e-23
tgo:TGME49_088350 nuclear NF-kB activating protein, putative 34.3 0.11
tgo:TGME49_021980 ww domain U1 zinc finger domain-containing p... 34.3 0.11
mmu:110876 Scn2a1, 6430408L10, A230052E19Rik, Nav1.2, Scn2a; s... 30.8 1.2
hsa:6326 SCN2A, BFIC3, EIEE11, HBA, HBSCI, HBSCII, NAC2, Na(v)... 30.8 1.2
pfa:PF13_0198 PfRh2a; reticulocyte binding protein 2 homolog A... 30.4 2.0
xla:100127307 fam166b; family with sequence similarity 166, me... 29.6 2.7
pfa:MAL13P1.176 PfRh2b; reticulocyte binding protein 2, homolog B 28.9 5.4
> tgo:TGME49_105240 XPA-binding protein, putative ; K12867 pre-mRNA-splicing
factor SYF1
Length=966
Score = 107 bits (267), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 57/125 (45%), Positives = 81/125 (64%), Gaps = 3/125 (2%)
Query 13 REEALQEQLRFQQKQQAEMQELEQAEAEFQELQRSRRQAATLASLQADAPLFVSASTFEG 72
RE+AL+EQL ++K++ + E+ QAE EF+ +Q R +A LASL DAP F++ S F+G
Sbjct 821 REKALEEQLYLKKKKELDDAEIAQAEEEFKRVQDMRARARMLASLPPDAPFFLAVSGFQG 880
Query 73 AHLGYAFKHGPQGLGYYLDEGQVGKGFIEARERSLLERSWPGGSGSSENAPPGLRRQDEE 132
GY F G QGLGYYLDE QVG F+ ARE++ + + PG +E G R + EE
Sbjct 881 VRPGYIFHRGAQGLGYYLDERQVGNDFVSAREKAFITDARPGARSRAEG---GDRERHEE 937
Query 133 MEIDI 137
++ID+
Sbjct 938 VDIDL 942
> tgo:TGME49_088350 nuclear NF-kB activating protein, putative
Length=903
Score = 34.3 bits (77), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 23/85 (27%), Positives = 39/85 (45%), Gaps = 9/85 (10%)
Query 13 REEALQEQLRFQQKQQAEMQELEQAEAEFQELQRSRRQAATLASLQADAPLFVSASTFEG 72
REE +Q ++Q K++ ++L +E + A +A FV +++F+G
Sbjct 534 REEKMQ---KWQAKEEKRRRKL------MKETKELEELELEEAWKKAGPKDFVKSASFKG 584
Query 73 AHLGYAFKHGPQGLGYYLDEGQVGK 97
G+ F G G GYY D + K
Sbjct 585 PRPGFVFHTGEAGTGYYKDRLSIKK 609
> tgo:TGME49_021980 ww domain U1 zinc finger domain-containing
protein
Length=591
Score = 34.3 bits (77), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 13/28 (46%), Positives = 17/28 (60%), Gaps = 0/28 (0%)
Query 64 FVSASTFEGAHLGYAFKHGPQGLGYYLD 91
F+ +S F G G+ F+ G GLGYY D
Sbjct 308 FLPSSVFAGKRAGFVFQLGSHGLGYYAD 335
> mmu:110876 Scn2a1, 6430408L10, A230052E19Rik, Nav1.2, Scn2a;
sodium channel, voltage-gated, type II, alpha 1; K04834 voltage-gated
sodium channel type II alpha
Length=2006
Score = 30.8 bits (68), Expect = 1.2, Method: Composition-based stats.
Identities = 10/24 (41%), Positives = 19/24 (79%), Gaps = 0/24 (0%)
Query 21 LRFQQKQQAEMQELEQAEAEFQEL 44
+ ++++ QA ++E EQ EAEFQ++
Sbjct 426 MAYEEQNQATLEEAEQKEAEFQQM 449
> hsa:6326 SCN2A, BFIC3, EIEE11, HBA, HBSCI, HBSCII, NAC2, Na(v)1.2,
Nav1.2, SCN2A1, SCN2A2; sodium channel, voltage-gated,
type II, alpha subunit; K04834 voltage-gated sodium channel
type II alpha
Length=2005
Score = 30.8 bits (68), Expect = 1.2, Method: Composition-based stats.
Identities = 10/24 (41%), Positives = 19/24 (79%), Gaps = 0/24 (0%)
Query 21 LRFQQKQQAEMQELEQAEAEFQEL 44
+ ++++ QA ++E EQ EAEFQ++
Sbjct 426 MAYEEQNQATLEEAEQKEAEFQQM 449
> pfa:PF13_0198 PfRh2a; reticulocyte binding protein 2 homolog
A; K13849 reticulocyte-binding protein
Length=3130
Score = 30.4 bits (67), Expect = 2.0, Method: Composition-based stats.
Identities = 20/64 (31%), Positives = 34/64 (53%), Gaps = 3/64 (4%)
Query 3 EDEERQQVLRREE-ALQEQLRFQQKQQAEMQELE--QAEAEFQELQRSRRQAATLASLQA 59
E E+++Q+ + EE QEQ R Q+++ + QE E Q E E + ++ R + LQ
Sbjct 2762 EREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQKEEELKRQEQERLEREKQEQLQK 2821
Query 60 DAPL 63
+ L
Sbjct 2822 EEEL 2825
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 32/49 (65%), Gaps = 2/49 (4%)
Query 3 EDEERQQVLRREE-ALQEQLRFQQKQQAEMQELEQAEAEFQELQRSRRQ 50
E E+++Q+ + EE QEQ R Q+++ + QE E+ + E +EL+R ++
Sbjct 2812 EREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQKE-EELKRQEQE 2859
Score = 28.9 bits (63), Expect = 4.6, Method: Composition-based stats.
Identities = 19/52 (36%), Positives = 31/52 (59%), Gaps = 10/52 (19%)
Query 2 KEDEERQQVLRR--------EEAL--QEQLRFQQKQQAEMQELEQAEAEFQE 43
+E ++++ L+R EEAL QEQ R Q++++ + QE E+ E E QE
Sbjct 2766 QEQLQKEEELKRQEQERLQKEEALKRQEQERLQKEEELKRQEQERLEREKQE 2817
> xla:100127307 fam166b; family with sequence similarity 166,
member B
Length=311
Score = 29.6 bits (65), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 27/109 (24%), Positives = 45/109 (41%), Gaps = 17/109 (15%)
Query 11 LRREEALQEQLRFQQKQQAEMQELEQAEAEFQELQRSRRQAATLASLQADAPLFVSASTF 70
E+ +QE R + +++QE Q + E ++ + R L +L + F+S F
Sbjct 129 FHNEQQMQESQRQEMLLTSKLQEGRQPQTEHEKQLLTARHRTPLPALSKEPAPFMSLRGF 188
Query 71 EGAHLGYAFKHGPQGLGYYLDEGQVGKGFIEA------RERSLLERSWP 113
+ PQG YY++E K FI R R L+ +P
Sbjct 189 Q-----------PQGSPYYMEEENPNKYFISGYTGYVPRSRFLIGNGYP 226
> pfa:MAL13P1.176 PfRh2b; reticulocyte binding protein 2, homolog
B
Length=3179
Score = 28.9 bits (63), Expect = 5.4, Method: Composition-based stats.
Identities = 15/48 (31%), Positives = 30/48 (62%), Gaps = 1/48 (2%)
Query 2 KEDEERQQVLRREEALQEQLRFQQKQQAEMQELEQAEAEFQELQRSRR 49
+E+ +RQ+ R E QEQL+ +++ + + QE +Q + QEL+ ++
Sbjct 2674 EEELKRQEQERLEREKQEQLQKEEELRKKEQE-KQQQRNIQELEEQKK 2720
Lambda K H
0.311 0.129 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2487377096
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40