bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0409_orf3
Length=106
Score E
Sequences producing significant alignments: (Bits) Value
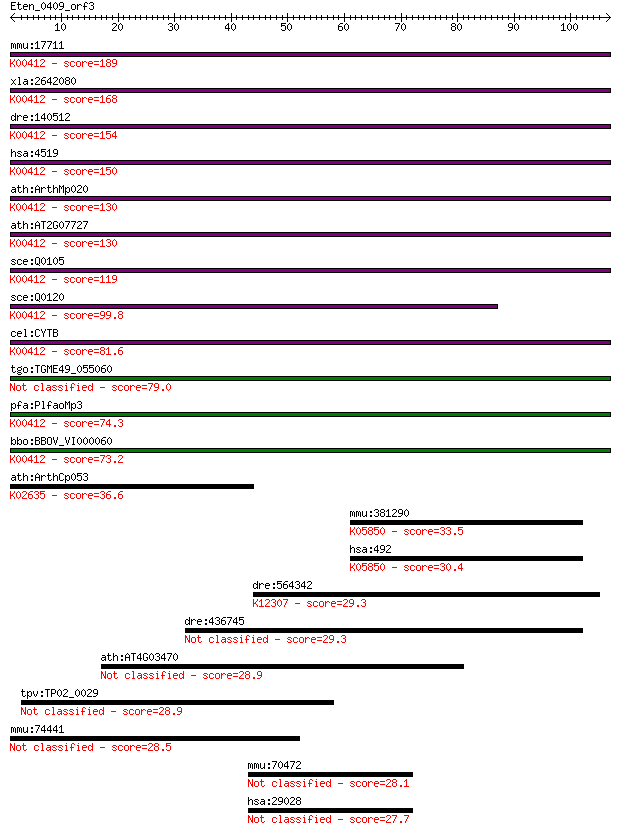
mmu:17711 CYTB; cytochrome b (EC:1.10.2.2); K00412 ubiquinol-c... 189 1e-48
xla:2642080 CYTB; cytochrome b; K00412 ubiquinol-cytochrome c ... 168 4e-42
dre:140512 CYTB, mtcyb; cytochrome b; K00412 ubiquinol-cytochr... 154 4e-38
hsa:4519 CYTB, MTCYB; cytochrome b; K00412 ubiquinol-cytochrom... 150 7e-37
ath:ArthMp020 cob; apocytochrome B; K00412 ubiquinol-cytochrom... 130 7e-31
ath:AT2G07727 cytochrome b (MTCYB) (COB) (CYTB); K00412 ubiqui... 130 7e-31
sce:Q0105 COB, COB1, CYTB; Cobp; K00412 ubiquinol-cytochrome c... 119 3e-27
sce:Q0120 BI4; Bi4p (EC:3.1.-.-); K00412 ubiquinol-cytochrome ... 99.8 2e-21
cel:CYTB cytochrome b; K00412 ubiquinol-cytochrome c reductase... 81.6 5e-16
tgo:TGME49_055060 cytochrome oxidase I (EC:1.9.3.1) 79.0 3e-15
pfa:PlfaoMp3 CYTB; cytochrome b; K00412 ubiquinol-cytochrome c... 74.3 9e-14
bbo:BBOV_VI000060 cytochrome b; K00412 ubiquinol-cytochrome c ... 73.2 2e-13
ath:ArthCp053 petB; cytochrome b6; K02635 cytochrome b6 36.6 0.019
mmu:381290 Atp2b4, MGC129362, MGC129363, PMCA4; ATPase, Ca++ t... 33.5 0.15
hsa:492 ATP2B3, PMCA3, PMCA3a; ATPase, Ca++ transporting, plas... 30.4 1.3
dre:564342 mfsd8, MGC136865, im:7139494, zgc:136865; major fac... 29.3 3.2
dre:436745 atp2b3a, atp2b3, pmca3a, zgc:92885; ATPase, Ca++ tr... 29.3 3.2
ath:AT4G03470 ankyrin repeat family protein 28.9 3.7
tpv:TP02_0029 hypothetical protein 28.9 3.9
mmu:74441 Slco6c1, 4933404A18Rik, C79179; solute carrier organ... 28.5 5.8
mmu:70472 Atad2, 2610509G12Rik, MGC38189; ATPase family, AAA d... 28.1 7.5
hsa:29028 ATAD2, ANCCA, DKFZp667N1320, MGC131938, MGC142216, M... 27.7 9.3
> mmu:17711 CYTB; cytochrome b (EC:1.10.2.2); K00412 ubiquinol-cytochrome
c reductase cytochrome b subunit [EC:1.10.2.2]
Length=381
Score = 189 bits (481), Expect = 1e-48, Method: Compositional matrix adjust.
Identities = 91/106 (85%), Positives = 98/106 (92%), Gaps = 0/106 (0%)
Query 1 GGFSVDKATLTRFFAFHFILPFIIIAIAIVHLLFLHETGSNNPTGISSDVDKIPFHPYYT 60
GGFSVDKATLTRFFAFHFILPFII A+AIVHLLFLHETGSNNPTG++SD DKIPFHPYYT
Sbjct 166 GGFSVDKATLTRFFAFHFILPFIIAALAIVHLLFLHETGSNNPTGLNSDADKIPFHPYYT 225
Query 61 IKDILGALLLILALILLVLFAPDLLGDPDNYTPANPLNTPPHIKPE 106
IKDILG L++ L L+ LVLF PD+LGDPDNY PANPLNTPPHIKPE
Sbjct 226 IKDILGILIMFLILMTLVLFFPDMLGDPDNYMPANPLNTPPHIKPE 271
> xla:2642080 CYTB; cytochrome b; K00412 ubiquinol-cytochrome
c reductase cytochrome b subunit [EC:1.10.2.2]
Length=379
Score = 168 bits (425), Expect = 4e-42, Method: Compositional matrix adjust.
Identities = 78/106 (73%), Positives = 94/106 (88%), Gaps = 0/106 (0%)
Query 1 GGFSVDKATLTRFFAFHFILPFIIIAIAIVHLLFLHETGSNNPTGISSDVDKIPFHPYYT 60
GGFSVD ATLTRFFAFHF+LPFII +I+HLLFLHETGS NPTG++SD DK+PFHPY++
Sbjct 167 GGFSVDNATLTRFFAFHFLLPFIIAGASILHLLFLHETGSTNPTGLNSDPDKVPFHPYFS 226
Query 61 IKDILGALLLILALILLVLFAPDLLGDPDNYTPANPLNTPPHIKPE 106
KD+LG L+++ AL LL +F+P+LLGDPDN+TPANPL TPPHIKPE
Sbjct 227 YKDLLGFLIMLTALTLLAMFSPNLLGDPDNFTPANPLITPPHIKPE 272
> dre:140512 CYTB, mtcyb; cytochrome b; K00412 ubiquinol-cytochrome
c reductase cytochrome b subunit [EC:1.10.2.2]
Length=380
Score = 154 bits (390), Expect = 4e-38, Method: Compositional matrix adjust.
Identities = 77/106 (72%), Positives = 96/106 (90%), Gaps = 0/106 (0%)
Query 1 GGFSVDKATLTRFFAFHFILPFIIIAIAIVHLLFLHETGSNNPTGISSDVDKIPFHPYYT 60
GGFSVD ATLTRFFAFHF+LPFIIIA+ I+HLLFLHETGSNNP G++ ++DKIPFHPY++
Sbjct 166 GGFSVDNATLTRFFAFHFLLPFIIIAMVILHLLFLHETGSNNPLGLNPNMDKIPFHPYFS 225
Query 61 IKDILGALLLILALILLVLFAPDLLGDPDNYTPANPLNTPPHIKPE 106
KD+LG ++++ +L LL LF+P+LLGDP+N+TPANPL TPPHIKPE
Sbjct 226 NKDLLGFVIMLFSLSLLALFSPNLLGDPENFTPANPLVTPPHIKPE 271
> hsa:4519 CYTB, MTCYB; cytochrome b; K00412 ubiquinol-cytochrome
c reductase cytochrome b subunit [EC:1.10.2.2]
Length=380
Score = 150 bits (380), Expect = 7e-37, Method: Compositional matrix adjust.
Identities = 83/106 (78%), Positives = 91/106 (85%), Gaps = 0/106 (0%)
Query 1 GGFSVDKATLTRFFAFHFILPFIIIAIAIVHLLFLHETGSNNPTGISSDVDKIPFHPYYT 60
GG+SVD TLTRFF FHFILPFII A+A +HLLFLHETGSNNP GI+S DKI FHPYYT
Sbjct 166 GGYSVDSPTLTRFFTFHFILPFIIAALATLHLLFLHETGSNNPLGITSHSDKITFHPYYT 225
Query 61 IKDILGALLLILALILLVLFAPDLLGDPDNYTPANPLNTPPHIKPE 106
IKD LG LL +L+L+ L LF+PDLLGDPDNYT ANPLNTPPHIKPE
Sbjct 226 IKDALGLLLFLLSLMTLTLFSPDLLGDPDNYTLANPLNTPPHIKPE 271
> ath:ArthMp020 cob; apocytochrome B; K00412 ubiquinol-cytochrome
c reductase cytochrome b subunit [EC:1.10.2.2]
Length=393
Score = 130 bits (328), Expect = 7e-31, Method: Compositional matrix adjust.
Identities = 56/106 (52%), Positives = 81/106 (76%), Gaps = 0/106 (0%)
Query 1 GGFSVDKATLTRFFAFHFILPFIIIAIAIVHLLFLHETGSNNPTGISSDVDKIPFHPYYT 60
GGFSVD ATL RFF+ H +LPFI++ +++HL LH+ GSNNP G+ S++DKI F+PY+
Sbjct 173 GGFSVDNATLNRFFSLHHLLPFILVGASLLHLAALHQYGSNNPLGVHSEMDKIAFYPYFY 232
Query 61 IKDILGALLLILALILLVLFAPDLLGDPDNYTPANPLNTPPHIKPE 106
+KD++G + + + + +AP++LG PDNY PANP++TPPHI PE
Sbjct 233 VKDLVGWVAFAIFFSIWIFYAPNVLGHPDNYIPANPMSTPPHIVPE 278
> ath:AT2G07727 cytochrome b (MTCYB) (COB) (CYTB); K00412 ubiquinol-cytochrome
c reductase cytochrome b subunit [EC:1.10.2.2]
Length=393
Score = 130 bits (328), Expect = 7e-31, Method: Compositional matrix adjust.
Identities = 56/106 (52%), Positives = 81/106 (76%), Gaps = 0/106 (0%)
Query 1 GGFSVDKATLTRFFAFHFILPFIIIAIAIVHLLFLHETGSNNPTGISSDVDKIPFHPYYT 60
GGFSVD ATL RFF+ H +LPFI++ +++HL LH+ GSNNP G+ S++DKI F+PY+
Sbjct 173 GGFSVDNATLNRFFSLHHLLPFILVGASLLHLAALHQYGSNNPLGVHSEMDKIAFYPYFY 232
Query 61 IKDILGALLLILALILLVLFAPDLLGDPDNYTPANPLNTPPHIKPE 106
+KD++G + + + + +AP++LG PDNY PANP++TPPHI PE
Sbjct 233 VKDLVGWVAFAIFFSIWIFYAPNVLGHPDNYIPANPMSTPPHIVPE 278
> sce:Q0105 COB, COB1, CYTB; Cobp; K00412 ubiquinol-cytochrome
c reductase cytochrome b subunit [EC:1.10.2.2]
Length=385
Score = 119 bits (297), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 55/106 (51%), Positives = 74/106 (69%), Gaps = 0/106 (0%)
Query 1 GGFSVDKATLTRFFAFHFILPFIIIAIAIVHLLFLHETGSNNPTGISSDVDKIPFHPYYT 60
GGFSV T+ RFFA H+++PFII A+ I+HL+ LH GS+NP GI+ ++D+IP H Y+
Sbjct 167 GGFSVSNPTIQRFFALHYLVPFIIAAMVIMHLMALHIHGSSNPLGITGNLDRIPMHSYFI 226
Query 61 IKDILGALLLILALILLVLFAPDLLGDPDNYTPANPLNTPPHIKPE 106
KD++ L +L L L V ++P+ LG PDNY P NPL TP I PE
Sbjct 227 FKDLVTVFLFMLILALFVFYSPNTLGHPDNYIPGNPLVTPASIVPE 272
> sce:Q0120 BI4; Bi4p (EC:3.1.-.-); K00412 ubiquinol-cytochrome
c reductase cytochrome b subunit [EC:1.10.2.2]
Length=638
Score = 99.8 bits (247), Expect = 2e-21, Method: Composition-based stats.
Identities = 42/86 (48%), Positives = 61/86 (70%), Gaps = 0/86 (0%)
Query 1 GGFSVDKATLTRFFAFHFILPFIIIAIAIVHLLFLHETGSNNPTGISSDVDKIPFHPYYT 60
GGFSV T+ RFFA H+++PFII A+ I+HL+ LH GS+NP GI+ ++D+IP H Y+
Sbjct 167 GGFSVSNPTIQRFFALHYLVPFIIAAMVIMHLMALHIHGSSNPLGITGNLDRIPMHSYFI 226
Query 61 IKDILGALLLILALILLVLFAPDLLG 86
KD++ L +L L L V ++P+ LG
Sbjct 227 FKDLVTVFLFMLILALFVFYSPNTLG 252
> cel:CYTB cytochrome b; K00412 ubiquinol-cytochrome c reductase
cytochrome b subunit [EC:1.10.2.2]
Length=370
Score = 81.6 bits (200), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 45/106 (42%), Positives = 61/106 (57%), Gaps = 1/106 (0%)
Query 1 GGFSVDKATLTRFFAFHFILPFIIIAIAIVHLLFLHETGSNNPTGISSDVDKIPFHPYYT 60
GF V ATL FF HF+LP+ I+ I + HL+FLH TGS + D DK+ F P Y
Sbjct 163 SGFGVTGATLKFFFVLHFLLPWAILVIVLGHLIFLHSTGSTSSLYCHGDYDKVCFSPEYL 222
Query 61 IKDILGALLLILALILLVLFAPDLLGDPDNYTPANPLNTPPHIKPE 106
KD ++ +L I+L L P LGD + + A+P+ +P HI PE
Sbjct 223 GKDAYNIVIWLL-FIVLSLIYPFNLGDAEMFIEADPMMSPVHIVPE 267
> tgo:TGME49_055060 cytochrome oxidase I (EC:1.9.3.1)
Length=402
Score = 79.0 bits (193), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 45/106 (42%), Positives = 58/106 (54%), Gaps = 1/106 (0%)
Query 1 GGFSVDKATLTRFFAFHFILPFIIIAIAIVHLLFLHETGSNNPTGISSDVDKIPFHPYYT 60
GG+ V TL RFF HFILPFI I ++H+ +LH GS+NP GI + + K+ F+P+
Sbjct 269 GGYYVSDVTLKRFFVLHFILPFIGCIIIVLHIFYLHLNGSSNPAGIDTAL-KVAFYPHML 327
Query 61 IKDILGALLLILALILLVLFAPDLLGDPDNYTPANPLNTPPHIKPE 106
+ D LI + L F L PDN P N TP HI PE
Sbjct 328 MTDAKCLSYLIGLIFLQAAFGLMELSHPDNSIPVNRFVTPLHIVPE 373
> pfa:PlfaoMp3 CYTB; cytochrome b; K00412 ubiquinol-cytochrome
c reductase cytochrome b subunit [EC:1.10.2.2]
Length=376
Score = 74.3 bits (181), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 44/106 (41%), Positives = 58/106 (54%), Gaps = 1/106 (0%)
Query 1 GGFSVDKATLTRFFAFHFILPFIIIAIAIVHLLFLHETGSNNPTGISSDVDKIPFHPYYT 60
GG++V T+ RFF HFILPFI + I +H+ FLH GS NP G + + KIPF+P
Sbjct 157 GGYTVSDPTIKRFFVLHFILPFIGLCIVFIHIFFLHLHGSTNPLGYDTAL-KIPFYPNLL 215
Query 61 IKDILGALLLILALILLVLFAPDLLGDPDNYTPANPLNTPPHIKPE 106
D+ G +I+ ++ LF L PDN N TP I PE
Sbjct 216 SLDVKGFNNVIILFLIQSLFGIIPLSHPDNAIVVNTYVTPSQIVPE 261
> bbo:BBOV_VI000060 cytochrome b; K00412 ubiquinol-cytochrome
c reductase cytochrome b subunit [EC:1.10.2.2]
Length=363
Score = 73.2 bits (178), Expect = 2e-13, Method: Composition-based stats.
Identities = 44/109 (40%), Positives = 62/109 (56%), Gaps = 4/109 (3%)
Query 1 GGFSVDKATLTRFFAFHFILPFIIIAIAIVHLLFLHETGSNNP-TGISSDVDKIPFHPYY 59
GG+ V TL RF+ HFILPF+++ + ++H+ +LH + S NP +G+ S + F+P
Sbjct 152 GGYGVAFPTLQRFYILHFILPFVLLIVVLIHIYYLHRSSSTNPLSGVDSWLVS-RFYPVI 210
Query 60 TIKDI--LGALLLILALILLVLFAPDLLGDPDNYTPANPLNTPPHIKPE 106
DI L L ++L + L P GD DN ANPL TP HI PE
Sbjct 211 IFSDIRMLTMLFILLGVQLTYGVIPLFQGDCDNSIMANPLQTPMHIVPE 259
> ath:ArthCp053 petB; cytochrome b6; K02635 cytochrome b6
Length=215
Score = 36.6 bits (83), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 27/44 (61%), Gaps = 1/44 (2%)
Query 1 GGFSVDKATLTRFFAFH-FILPFIIIAIAIVHLLFLHETGSNNP 43
G SV ++TLTRF++ H F+LP + ++H L + + G + P
Sbjct 171 GSASVGQSTLTRFYSLHTFVLPLLTAVFMLMHFLMIRKQGISGP 214
> mmu:381290 Atp2b4, MGC129362, MGC129363, PMCA4; ATPase, Ca++
transporting, plasma membrane 4 (EC:3.6.3.8); K05850 Ca2+ transporting
ATPase, plasma membrane [EC:3.6.3.8]
Length=1205
Score = 33.5 bits (75), Expect = 0.15, Method: Composition-based stats.
Identities = 18/41 (43%), Positives = 27/41 (65%), Gaps = 1/41 (2%)
Query 61 IKDILGALLLILALILLVLFAPDLLGDPDNYTPANPLNTPP 101
+K+ILG + L ++ L++FA D L D D+ A PLN+PP
Sbjct 918 MKNILGHAVYQLLIVFLLVFAGDTLFDIDSGRKA-PLNSPP 957
> hsa:492 ATP2B3, PMCA3, PMCA3a; ATPase, Ca++ transporting, plasma
membrane 3 (EC:3.6.3.8); K05850 Ca2+ transporting ATPase,
plasma membrane [EC:3.6.3.8]
Length=1220
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 26/41 (63%), Gaps = 1/41 (2%)
Query 61 IKDILGALLLILALILLVLFAPDLLGDPDNYTPANPLNTPP 101
+K+ILG + LA+I +LF +L D D+ A PL++PP
Sbjct 926 MKNILGHAVYQLAIIFTLLFVGELFFDIDSGRNA-PLHSPP 965
> dre:564342 mfsd8, MGC136865, im:7139494, zgc:136865; major facilitator
superfamily domain containing 8; K12307 MFS transporter,
ceroid-lipofuscinosis neuronal protein 7
Length=504
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 20/68 (29%), Positives = 35/68 (51%), Gaps = 8/68 (11%)
Query 44 TGISSDVDKIPFHPYYTIKDILGALLLILALILLVLFAPDLLGD-------PDNYTPANP 96
TG+S DV K+ + YT +L A ++ ++L++L + D P NYTP
Sbjct 188 TGVSVDVIKLQVN-MYTAPALLAACFGVINILLVILVLREHSVDDHGNRIRPINYTPEER 246
Query 97 LNTPPHIK 104
++ PP ++
Sbjct 247 VDVPPEVE 254
> dre:436745 atp2b3a, atp2b3, pmca3a, zgc:92885; ATPase, Ca++
transporting, plasma membrane 3a
Length=1176
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 24/70 (34%), Positives = 35/70 (50%), Gaps = 12/70 (17%)
Query 32 LLFLHETGSNNPTGISSDVDKIPFHPYYTIKDILGALLLILALILLVLFAPDLLGDPDNY 91
LL G NNP IS + +K+ILG + L +I +LFA + + D D+
Sbjct 908 LLLRKPYGRNNPL-ISRTM----------MKNILGHAVYQLVIIFTLLFAGERIFDIDSG 956
Query 92 TPANPLNTPP 101
A PL++PP
Sbjct 957 RDA-PLHSPP 965
> ath:AT4G03470 ankyrin repeat family protein
Length=683
Score = 28.9 bits (63), Expect = 3.7, Method: Composition-based stats.
Identities = 20/64 (31%), Positives = 30/64 (46%), Gaps = 5/64 (7%)
Query 17 HFILPFIIIAIAIVHLLFLHETGSNNPTGISSDVDKIPFHPYYTIKDILGALLLILALIL 76
H+I + +A+V L F N+P + + P P KD + ALL++ ALI
Sbjct 454 HYIFRERLTLLALVQLHF-----QNDPRCAHTMIQTRPIMPQGGNKDYINALLVVAALIT 508
Query 77 LVLF 80
V F
Sbjct 509 TVTF 512
> tpv:TP02_0029 hypothetical protein
Length=1314
Score = 28.9 bits (63), Expect = 3.9, Method: Composition-based stats.
Identities = 16/55 (29%), Positives = 28/55 (50%), Gaps = 3/55 (5%)
Query 3 FSVDKATLTRFFAFHFILPFIIIAIAIVHLLFLHETGSNNPTGISSDVDKIPFHP 57
F+V ++ L + ++ P ++ I H + E+ N+P ISS +D PF P
Sbjct 134 FTVIQSPLDKLVEYN---PDLMAQINSDHFILEEESDLNDPKSISSSIDLSPFLP 185
> mmu:74441 Slco6c1, 4933404A18Rik, C79179; solute carrier organic
anion transporter family, member 6c1
Length=706
Score = 28.5 bits (62), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 14/51 (27%), Positives = 24/51 (47%), Gaps = 0/51 (0%)
Query 1 GGFSVDKATLTRFFAFHFILPFIIIAIAIVHLLFLHETGSNNPTGISSDVD 51
GG VD+ +T F L ++++ + L+F E + GI+ D D
Sbjct 429 GGLIVDRLEMTNKNKLKFTLVTTVVSVGLFLLIFFVECQTTTFAGINEDYD 479
> mmu:70472 Atad2, 2610509G12Rik, MGC38189; ATPase family, AAA
domain containing 2 (EC:3.6.1.3)
Length=1364
Score = 28.1 bits (61), Expect = 7.5, Method: Composition-based stats.
Identities = 13/29 (44%), Positives = 17/29 (58%), Gaps = 0/29 (0%)
Query 43 PTGISSDVDKIPFHPYYTIKDILGALLLI 71
P +SS + KI H Y T+KD L + LI
Sbjct 1001 PMDLSSVISKIDLHKYLTVKDYLKDIDLI 1029
> hsa:29028 ATAD2, ANCCA, DKFZp667N1320, MGC131938, MGC142216,
MGC29843, MGC5254, PRO2000; ATPase family, AAA domain containing
2 (EC:3.6.1.3)
Length=1390
Score = 27.7 bits (60), Expect = 9.3, Method: Composition-based stats.
Identities = 13/29 (44%), Positives = 17/29 (58%), Gaps = 0/29 (0%)
Query 43 PTGISSDVDKIPFHPYYTIKDILGALLLI 71
P +SS + KI H Y T+KD L + LI
Sbjct 1028 PMDLSSVISKIDLHKYLTVKDYLRDIDLI 1056
Lambda K H
0.327 0.149 0.457
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2012750684
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40