bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0414_orf2
Length=169
Score E
Sequences producing significant alignments: (Bits) Value
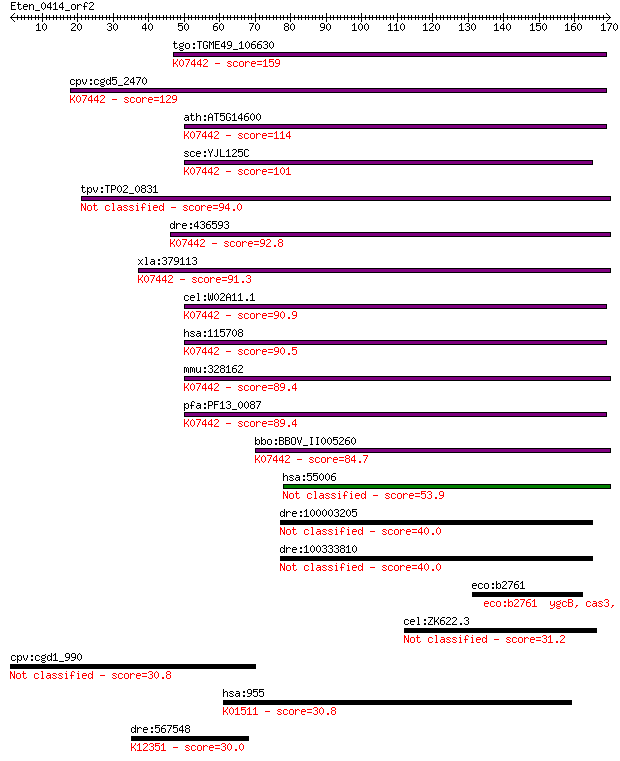
tgo:TGME49_106630 tRNA (adenine-N(1)-)-methyltransferase catal... 159 4e-39
cpv:cgd5_2470 GCD14 RNA methylase ; K07442 tRNA (adenine-N1-)-... 129 4e-30
ath:AT5G14600 tRNA (adenine-N1-)-methyltransferase; K07442 tRN... 114 1e-25
sce:YJL125C GCD14, TRM61; 1-methyladenosine tRNA methyltransfe... 101 1e-21
tpv:TP02_0831 hypothetical protein 94.0 2e-19
dre:436593 trmt61a, zgc:86657; tRNA methyltransferase 61 homol... 92.8 5e-19
xla:379113 trmt61a, MGC53312; tRNA methyltransferase 61 homolo... 91.3 1e-18
cel:W02A11.1 hypothetical protein; K07442 tRNA (adenine-N1-)-m... 90.9 2e-18
hsa:115708 TRMT61A, C14orf172, FLJ40452, GCD14, Gcd14p, TRM61,... 90.5 3e-18
mmu:328162 Trmt61a, 6720458F09Rik, AI606093, Gcd14, Trm61; tRN... 89.4 5e-18
pfa:PF13_0087 methyltransferase, putative; K07442 tRNA (adenin... 89.4 6e-18
bbo:BBOV_II005260 18.m06436; hypothetical protein; K07442 tRNA... 84.7 1e-16
hsa:55006 TRMT61B, DKFZp564I2178, FLJ20628; tRNA methyltransfe... 53.9 2e-07
dre:100003205 hypothetical LOC100003205 40.0 0.003
dre:100333810 hypothetical protein LOC100333810 40.0 0.004
eco:b2761 ygcB, cas3, ECK2756, JW2731; Cas3 predicted helicase... 31.2 2.0
cel:ZK622.3 pmt-1; Phosphoethanolamine MethyTransferase family... 31.2 2.0
cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alph... 30.8 2.0
hsa:955 ENTPD6, CD39L2, DKFZp781G2277, DKFZp781K21102, FLJ3671... 30.8 2.2
dre:567548 smpd2, NSMase; sphingomyelin phosphodiesterase 2, n... 30.0 3.7
> tgo:TGME49_106630 tRNA (adenine-N(1)-)-methyltransferase catalytic
subunit, putative (EC:2.1.1.36); K07442 tRNA (adenine-N1-)-methyltransferase
catalytic subunit [EC:2.1.1.36]
Length=360
Score = 159 bits (402), Expect = 4e-39, Method: Compositional matrix adjust.
Identities = 71/122 (58%), Positives = 94/122 (77%), Gaps = 0/122 (0%)
Query 47 NFPKEGDLVLLYGSRDIIVPCVLRKGGVCRTRLGNFEHADIMRTPMGQKVQDRRSGKWLV 106
+P+ GD V+LYGS +++P +L +G V +RLGN+ H DI+ T G KVQDR+S +WLV
Sbjct 76 EYPRYGDFVILYGSHQLVIPVLLERGRVSNSRLGNYRHDDIVTTAYGSKVQDRKSKRWLV 135
Query 107 VLRPTPDLHTLALRHRTQIIYHADISLIMSLLDVCPGKTIIEAGTGSGSLTVSLAASLRP 166
VLRP+PDL +LAL HRTQI+YH+DISL++ LLD CPG+ I EAGTGSGSL+ LA ++ P
Sbjct 136 VLRPSPDLFSLALTHRTQILYHSDISLVLMLLDACPGRRICEAGTGSGSLSCHLARAVAP 195
Query 167 GG 168
G
Sbjct 196 TG 197
> cpv:cgd5_2470 GCD14 RNA methylase ; K07442 tRNA (adenine-N1-)-methyltransferase
catalytic subunit [EC:2.1.1.36]
Length=312
Score = 129 bits (325), Expect = 4e-30, Method: Compositional matrix adjust.
Identities = 73/161 (45%), Positives = 95/161 (59%), Gaps = 10/161 (6%)
Query 18 EGAPGPCI---VEESGPDLPEHFTKEQDLYPS-------NFPKEGDLVLLYGSRDIIVPC 67
EG P P V S DL E+F + S K GDLV+L+G ++++
Sbjct 5 EGIPEPTNLGNVSASLEDLEENFEGSNIVNKSYTQRRRYEMCKNGDLVVLFGGYEMVIQV 64
Query 68 VLRKGGVCRTRLGNFEHADIMRTPMGQKVQDRRSGKWLVVLRPTPDLHTLALRHRTQIIY 127
L + G +TR G F H +I+ G ++ D KW+VVL+ +P+L +++L HRTQI+Y
Sbjct 65 FLEQNGRTQTRNGVFLHNEIIGKNFGSRIFDTSKSKWMVVLKLSPELISISLNHRTQILY 124
Query 128 HADISLIMSLLDVCPGKTIIEAGTGSGSLTVSLAASLRPGG 168
ADISLI LLD PGK IIEAGTGSGSLTVSL S PGG
Sbjct 125 QADISLICVLLDASPGKNIIEAGTGSGSLTVSLCRSTNPGG 165
> ath:AT5G14600 tRNA (adenine-N1-)-methyltransferase; K07442 tRNA
(adenine-N1-)-methyltransferase catalytic subunit [EC:2.1.1.36]
Length=318
Score = 114 bits (285), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 55/119 (46%), Positives = 83/119 (69%), Gaps = 1/119 (0%)
Query 50 KEGDLVLLYGSRDIIVPCVLRKGGVCRTRLGNFEHADIMRTPMGQKVQDRRSGKWLVVLR 109
++GDLV++Y D++ P + K GV + R G ++H+D + P+G KV + GK++ +L
Sbjct 17 EDGDLVIVYERHDVMKPVKVSKDGVLQNRFGVYKHSDWIGKPLGTKVFSNK-GKFVYLLA 75
Query 110 PTPDLHTLALRHRTQIIYHADISLIMSLLDVCPGKTIIEAGTGSGSLTVSLAASLRPGG 168
PTP+L TL L HRTQI+Y ADIS ++ L+V PG ++E+GTGSGSL+ SLA ++ P G
Sbjct 76 PTPELWTLVLSHRTQILYIADISFVVMYLEVVPGCVVLESGTGSGSLSTSLARAVAPTG 134
> sce:YJL125C GCD14, TRM61; 1-methyladenosine tRNA methyltransferase
subunit (EC:2.1.1.36); K07442 tRNA (adenine-N1-)-methyltransferase
catalytic subunit [EC:2.1.1.36]
Length=383
Score = 101 bits (252), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 55/119 (46%), Positives = 73/119 (61%), Gaps = 4/119 (3%)
Query 50 KEGDLVLLYGSRDIIVPCVLRKGGVCRTRLGNFEHADIMRTPMGQKVQDRRSGK----WL 105
KEGDL L++ SRD I P + V TR G+F H DI+ P G ++ R G ++
Sbjct 14 KEGDLTLIWVSRDNIKPVRMHSEEVFNTRYGSFPHKDIIGKPYGSQIAIRTKGSNKFAFV 73
Query 106 VVLRPTPDLHTLALRHRTQIIYHADISLIMSLLDVCPGKTIIEAGTGSGSLTVSLAASL 164
VL+PTP+L TL+L HRTQI+Y D S IM L+ P +IEAGTGSGS + + A S+
Sbjct 74 HVLQPTPELWTLSLPHRTQIVYTPDSSYIMQRLNCSPHSRVIEAGTGSGSFSHAFARSV 132
> tpv:TP02_0831 hypothetical protein
Length=312
Score = 94.0 bits (232), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 60/156 (38%), Positives = 86/156 (55%), Gaps = 26/156 (16%)
Query 21 PGPCIVEESGPDLPEHFTKEQDLYPSNFPKEGDLVLLYGSRDIIVPCVLRKGGVCRTRL- 79
PG ++ SGP+ ++Y S PKE D +P + V RL
Sbjct 5 PGDPVILFSGPN---------NIYFSQIPKE----------DQKIPNKQSEANVKNKRLI 45
Query 80 ----GNFEHADIMRTPMGQKV--QDRRSGKWLVVLRPTPDLHTLALRHRTQIIYHADISL 133
G F+ A + GQK+ + W+V+L+PTP+L T ++ HRTQI+Y ADISL
Sbjct 46 HNKKGIFDLASCIGKFYGQKLFWDENNKKHWVVLLKPTPELITKSIIHRTQILYRADISL 105
Query 134 IMSLLDVCPGKTIIEAGTGSGSLTVSLAASLRPGGR 169
I+ LLD+ PGK ++E GTGSGSL+ +LA+S+ P G
Sbjct 106 IVLLLDLMPGKRVLECGTGSGSLSYALASSVAPNGH 141
> dre:436593 trmt61a, zgc:86657; tRNA methyltransferase 61 homolog
A (S. cerevisiae) (EC:2.1.1.36); K07442 tRNA (adenine-N1-)-methyltransferase
catalytic subunit [EC:2.1.1.36]
Length=305
Score = 92.8 bits (229), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 49/125 (39%), Positives = 77/125 (61%), Gaps = 2/125 (1%)
Query 46 SNFPKEGDLVLLYGSRDIIVPCVLRKGGVCRTRLGNFEHA-DIMRTPMGQKVQDRRSGKW 104
S +EGD+V+++ D ++P ++ G +TR G H+ D++ G KV + G W
Sbjct 7 SELIQEGDVVIIFMGHDSMMPIKVQSGAQTQTRYGVIRHSSDLIGQRFGSKVTCSKGG-W 65
Query 105 LVVLRPTPDLHTLALRHRTQIIYHADISLIMSLLDVCPGKTIIEAGTGSGSLTVSLAASL 164
+ VL PTP+L T+ L HRTQI+Y DI+ I +L++ PG + E+GTGSGSL+ S+ ++
Sbjct 66 VYVLHPTPELWTINLPHRTQILYTTDIANITLMLELKPGSVVCESGTGSGSLSHSILRTI 125
Query 165 RPGGR 169
P G
Sbjct 126 APTGH 130
> xla:379113 trmt61a, MGC53312; tRNA methyltransferase 61 homolog
A; K07442 tRNA (adenine-N1-)-methyltransferase catalytic
subunit [EC:2.1.1.36]
Length=303
Score = 91.3 bits (225), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 49/134 (36%), Positives = 79/134 (58%), Gaps = 7/134 (5%)
Query 37 FTKEQDLYPSNFPKEGDLVLLYGSRDIIVPCVLRKGGVCRTRLGNFEHA-DIMRTPMGQK 95
F + +DL ++GD +++ D + P ++ G +T+ G H+ D++ G K
Sbjct 3 FVEYKDLI-----EDGDTAIVFLGHDSMFPITIQNGSQTQTKYGVIRHSTDLIGKKFGSK 57
Query 96 VQDRRSGKWLVVLRPTPDLHTLALRHRTQIIYHADISLIMSLLDVCPGKTIIEAGTGSGS 155
V + G W+ +L PTP+L TL L HRTQI+Y DISLI +L++ PG + E+GTGSGS
Sbjct 58 VTCSKGG-WVYILYPTPELWTLNLPHRTQILYSTDISLITMMLELKPGSVVCESGTGSGS 116
Query 156 LTVSLAASLRPGGR 169
L+ ++ ++ P G
Sbjct 117 LSHAIIRTVAPTGH 130
> cel:W02A11.1 hypothetical protein; K07442 tRNA (adenine-N1-)-methyltransferase
catalytic subunit [EC:2.1.1.36]
Length=312
Score = 90.9 bits (224), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 48/119 (40%), Positives = 74/119 (62%), Gaps = 2/119 (1%)
Query 50 KEGDLVLLYGSRDIIVPCVLRKGGVCRTRLGNFEHADIMRTPMGQKVQDRRSGKWLVVLR 109
+EGD V++Y + +VP ++++G + G H I+ GQ++ + ++ +LR
Sbjct 16 QEGDTVIVYVTYGQMVPTIVKRGQTLMMKYGALRHEFIIGKRWGQRLSA--TAGYVYILR 73
Query 110 PTPDLHTLALRHRTQIIYHADISLIMSLLDVCPGKTIIEAGTGSGSLTVSLAASLRPGG 168
PT DL T AL RTQI+Y AD + I+SLLD PG I E+GTGSGSL+ ++A ++ P G
Sbjct 74 PTSDLWTRALPRRTQILYTADCAQILSLLDAKPGSVICESGTGSGSLSHAIALTVAPTG 132
> hsa:115708 TRMT61A, C14orf172, FLJ40452, GCD14, Gcd14p, TRM61,
hTRM61; tRNA methyltransferase 61 homolog A (S. cerevisiae)
(EC:2.1.1.36); K07442 tRNA (adenine-N1-)-methyltransferase
catalytic subunit [EC:2.1.1.36]
Length=289
Score = 90.5 bits (223), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 50/120 (41%), Positives = 74/120 (61%), Gaps = 2/120 (1%)
Query 50 KEGDLVLLYGSRDIIVPCVLRKGGVCRTRLGNFEHA-DIMRTPMGQKVQDRRSGKWLVVL 108
KEGD +L +V +++G +TR G H+ D++ P G KV R G W+ VL
Sbjct 11 KEGDTAILSLGHGAMVAVRVQRGAQTQTRHGVLRHSVDLIGRPFGSKVTCGRGG-WVYVL 69
Query 109 RPTPDLHTLALRHRTQIIYHADISLIMSLLDVCPGKTIIEAGTGSGSLTVSLAASLRPGG 168
PTP+L TL L HRTQI+Y DI+LI +L++ PG + E+GTGSGS++ ++ ++ P G
Sbjct 70 HPTPELWTLNLPHRTQILYSTDIALITMMLELRPGSVVCESGTGSGSVSHAIIRTIAPTG 129
> mmu:328162 Trmt61a, 6720458F09Rik, AI606093, Gcd14, Trm61; tRNA
methyltransferase 61 homolog A (S. cerevisiae) (EC:2.1.1.36);
K07442 tRNA (adenine-N1-)-methyltransferase catalytic
subunit [EC:2.1.1.36]
Length=290
Score = 89.4 bits (220), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 50/121 (41%), Positives = 74/121 (61%), Gaps = 2/121 (1%)
Query 50 KEGDLVLLYGSRDIIVPCVLRKGGVCRTRLGNFEHA-DIMRTPMGQKVQDRRSGKWLVVL 108
KEGD +L +V +++G +TR G H+ D++ P G KV R G W+ VL
Sbjct 11 KEGDTAILSLGHGSMVAVRVQRGAQTQTRHGVLRHSVDLIGRPFGSKVICSRGG-WVYVL 69
Query 109 RPTPDLHTLALRHRTQIIYHADISLIMSLLDVCPGKTIIEAGTGSGSLTVSLAASLRPGG 168
PTP+L T+ L HRTQI+Y DI+LI +L++ PG + E+GTGSGS++ ++ S+ P G
Sbjct 70 HPTPELWTVNLPHRTQILYSTDIALITMMLELRPGSVVCESGTGSGSVSHAIIRSVAPTG 129
Query 169 R 169
Sbjct 130 H 130
> pfa:PF13_0087 methyltransferase, putative; K07442 tRNA (adenine-N1-)-methyltransferase
catalytic subunit [EC:2.1.1.36]
Length=316
Score = 89.4 bits (220), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 44/119 (36%), Positives = 67/119 (56%), Gaps = 0/119 (0%)
Query 50 KEGDLVLLYGSRDIIVPCVLRKGGVCRTRLGNFEHADIMRTPMGQKVQDRRSGKWLVVLR 109
KEG+ V++Y + I L G+ + G F H DI+ G K+ D + ++ VL+
Sbjct 3 KEGNCVVIYSDEENIDLLKLTNDGIINNKKGRFLHKDIIGKEYGSKIYDTLNKNYIYVLK 62
Query 110 PTPDLHTLALRHRTQIIYHADISLIMSLLDVCPGKTIIEAGTGSGSLTVSLAASLRPGG 168
+P+L L+ +TQ +Y DIS I + + P K I+EAGTG+G LT +LA S+ P G
Sbjct 63 RSPELIVNCLKKKTQTLYEHDISFICLVCNALPNKKILEAGTGTGCLTYALANSVLPNG 121
> bbo:BBOV_II005260 18.m06436; hypothetical protein; K07442 tRNA
(adenine-N1-)-methyltransferase catalytic subunit [EC:2.1.1.36]
Length=286
Score = 84.7 bits (208), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 46/101 (45%), Positives = 64/101 (63%), Gaps = 8/101 (7%)
Query 70 RKGGVCRTRLGNFEHADIMRTPMGQKVQ-DRRSGKWLVVLRPTPDLHTLALRHRTQIIYH 128
RKGG+ F+ ++ GQK D + +W V+L+PT +L T + HRTQI+Y
Sbjct 47 RKGGI-------FDLRKVVGKQYGQKFYWDSVANRWSVILQPTGELITRTVTHRTQILYR 99
Query 129 ADISLIMSLLDVCPGKTIIEAGTGSGSLTVSLAASLRPGGR 169
ADISL + LLD+ PGK ++E GTGSGSL+ SLA ++ P G
Sbjct 100 ADISLAILLLDLYPGKKVLECGTGSGSLSYSLATTVAPTGH 140
> hsa:55006 TRMT61B, DKFZp564I2178, FLJ20628; tRNA methyltransferase
61 homolog B (S. cerevisiae) (EC:2.1.1.36)
Length=477
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 33/101 (32%), Positives = 60/101 (59%), Gaps = 9/101 (8%)
Query 78 RLGNFE--HADIMRTPMGQKV-----QDRRS--GKWLVVLRPTPDLHTLALRHRTQIIYH 128
RL NF +++ P G+ V Q RS GK ++ RP + + + ++ T I +
Sbjct 173 RLNNFGLLNSNWGAVPFGKIVGKFPGQILRSSFGKQYMLRRPALEDYVVLMKRGTAITFP 232
Query 129 ADISLIMSLLDVCPGKTIIEAGTGSGSLTVSLAASLRPGGR 169
DI++I+S++D+ PG T++EAG+GSG +++ L+ ++ GR
Sbjct 233 KDINMILSMMDINPGDTVLEAGSGSGGMSLFLSKAVGSQGR 273
> dre:100003205 hypothetical LOC100003205
Length=450
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 22/88 (25%), Positives = 47/88 (53%), Gaps = 1/88 (1%)
Query 77 TRLGNFEHADIMRTPMGQKVQDRRSGKWLVVLRPTPDLHTLALRHRTQIIYHADISLIMS 136
+ G+ H D+ P G ++ + G V+ R + + L ++ I Y D S +++
Sbjct 168 SNWGSVAHDDMEGRPAGSVIRTSK-GIPFVIRRVSLEEFVLFMKRGPAIAYPKDASAMLT 226
Query 137 LLDVCPGKTIIEAGTGSGSLTVSLAASL 164
++DV G ++E G+GSG +++ L+ ++
Sbjct 227 MMDVTEGDCVLECGSGSGGMSLFLSRAV 254
> dre:100333810 hypothetical protein LOC100333810
Length=445
Score = 40.0 bits (92), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 22/88 (25%), Positives = 47/88 (53%), Gaps = 1/88 (1%)
Query 77 TRLGNFEHADIMRTPMGQKVQDRRSGKWLVVLRPTPDLHTLALRHRTQIIYHADISLIMS 136
+ G+ H D+ P G ++ + G V+ R + + L ++ I Y D S +++
Sbjct 163 SNWGSVAHDDMEGRPAGSVIRTSK-GIPFVIRRVSLEEFVLFMKRGPAIAYPKDASAMLT 221
Query 137 LLDVCPGKTIIEAGTGSGSLTVSLAASL 164
++DV G ++E G+GSG +++ L+ ++
Sbjct 222 MMDVTKGDCVLECGSGSGGMSLFLSRAV 249
> eco:b2761 ygcB, cas3, ECK2756, JW2731; Cas3 predicted helicase
needed for Cascade anti-viral activity; K07012
Length=888
Score = 31.2 bits (69), Expect = 2.0, Method: Composition-based stats.
Identities = 14/31 (45%), Positives = 20/31 (64%), Gaps = 0/31 (0%)
Query 131 ISLIMSLLDVCPGKTIIEAGTGSGSLTVSLA 161
+ +++ L V PG T+IEA TGSG +LA
Sbjct 296 LQVLVDALPVAPGLTVIEAPTGSGKTETALA 326
> cel:ZK622.3 pmt-1; Phosphoethanolamine MethyTransferase family
member (pmt-1)
Length=475
Score = 31.2 bits (69), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 15/54 (27%), Positives = 29/54 (53%), Gaps = 0/54 (0%)
Query 112 PDLHTLALRHRTQIIYHADISLIMSLLDVCPGKTIIEAGTGSGSLTVSLAASLR 165
PD +++ L H + + +D + I++ L + K +++ G G G T LA + R
Sbjct 35 PDTNSMMLNHSAEELESSDRADILASLPLLHNKDVVDIGAGIGRFTTVLAETAR 88
> cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alpha
plus beta subunits, duplicated gene, 12 transmembrane domain
(EC:1.6.1.2)
Length=1147
Score = 30.8 bits (68), Expect = 2.0, Method: Composition-based stats.
Identities = 24/69 (34%), Positives = 34/69 (49%), Gaps = 3/69 (4%)
Query 1 EARGSIVLINMARRATGEGAPGPCIVEESGPDLPEHFTKEQDLYPSNFPKEGDLVLLYGS 60
E+R +IV I + A ++ ESG +P KE D +N E DLVL+ G+
Sbjct 385 ESRSTIVEIGIHPVAGRMPGHMNVLLAESG--VPSRIVKEMDAV-NNCMHEYDLVLVVGA 441
Query 61 RDIIVPCVL 69
DI+ P L
Sbjct 442 NDIVNPAAL 450
> hsa:955 ENTPD6, CD39L2, DKFZp781G2277, DKFZp781K21102, FLJ36711,
IL-6SAG, IL6ST2, NTPDase-6, dJ738P15.3; ectonucleoside
triphosphate diphosphohydrolase 6 (putative) (EC:3.6.1.6); K01511
ectonucleoside triphosphate diphosphohydrolase 5/6 [EC:3.6.1.6]
Length=467
Score = 30.8 bits (68), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 27/98 (27%), Positives = 42/98 (42%), Gaps = 11/98 (11%)
Query 61 RDIIVPCVLRKGGVCRTRLGNFEHADIMRTPMGQKVQDRRSGKWLVVLRPTPDLHTLALR 120
++++ PC+ + G +EHA++ GQK S L R + L
Sbjct 302 KELVSPCL------SPSFKGEWEHAEVTYRVSGQKAA--ASLHELCAARVSEVLQNRV-- 351
Query 121 HRTQIIYHADISLIMSLLDVCPGKTIIEAGTGSGSLTV 158
HRT+ + H D D+ G +I+A G GSL V
Sbjct 352 HRTEEVKHVDFYAFSYYYDLAAGVGLIDAEKG-GSLVV 388
> dre:567548 smpd2, NSMase; sphingomyelin phosphodiesterase 2,
neutral (EC:3.1.4.12); K12351 sphingomyelin phosphodiesterase
2 [EC:3.1.4.12]
Length=420
Score = 30.0 bits (66), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 14/33 (42%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 35 EHFTKEQDLYPSNFPKEGDLVLLYGSRDIIVPC 67
HFTK+QDL P D +L+ GS+ + V C
Sbjct 220 NHFTKKQDLIPFEKGIRIDYILMKGSQRVSVKC 252
Lambda K H
0.321 0.140 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4222647260
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40