bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0415_orf3
Length=249
Score E
Sequences producing significant alignments: (Bits) Value
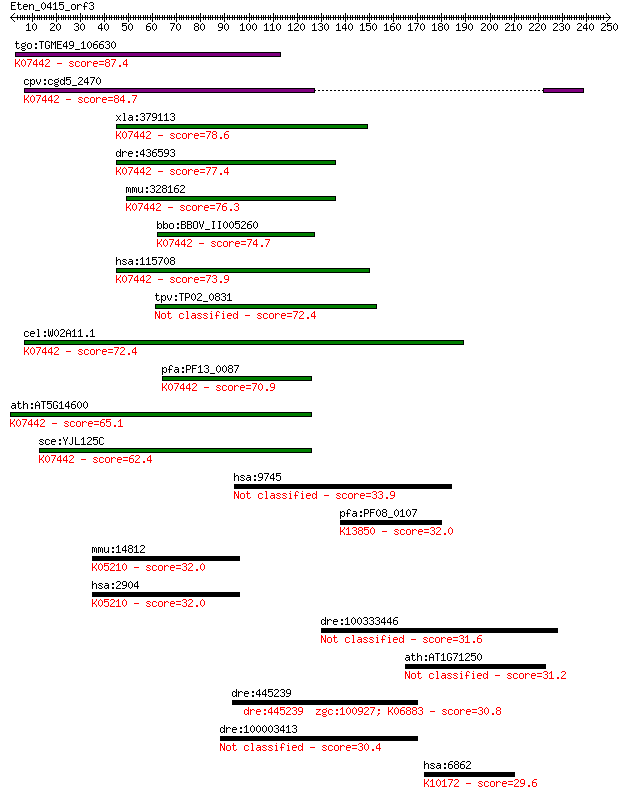
tgo:TGME49_106630 tRNA (adenine-N(1)-)-methyltransferase catal... 87.4 4e-17
cpv:cgd5_2470 GCD14 RNA methylase ; K07442 tRNA (adenine-N1-)-... 84.7 3e-16
xla:379113 trmt61a, MGC53312; tRNA methyltransferase 61 homolo... 78.6 2e-14
dre:436593 trmt61a, zgc:86657; tRNA methyltransferase 61 homol... 77.4 4e-14
mmu:328162 Trmt61a, 6720458F09Rik, AI606093, Gcd14, Trm61; tRN... 76.3 9e-14
bbo:BBOV_II005260 18.m06436; hypothetical protein; K07442 tRNA... 74.7 3e-13
hsa:115708 TRMT61A, C14orf172, FLJ40452, GCD14, Gcd14p, TRM61,... 73.9 5e-13
tpv:TP02_0831 hypothetical protein 72.4 1e-12
cel:W02A11.1 hypothetical protein; K07442 tRNA (adenine-N1-)-m... 72.4 1e-12
pfa:PF13_0087 methyltransferase, putative; K07442 tRNA (adenin... 70.9 4e-12
ath:AT5G14600 tRNA (adenine-N1-)-methyltransferase; K07442 tRN... 65.1 2e-10
sce:YJL125C GCD14, TRM61; 1-methyladenosine tRNA methyltransfe... 62.4 1e-09
hsa:9745 ZNF536, KIAA0390; zinc finger protein 536 33.9
pfa:PF08_0107 VAR; erythrocyte membrane protein 1, PfEMP1; K13... 32.0 1.9
mmu:14812 Grin2b, AW490526, NR2B, Nmdar2b; glutamate receptor,... 32.0 2.0
hsa:2904 GRIN2B, MGC142178, MGC142180, NMDAR2B, NR2B, hNR3; gl... 32.0 2.0
dre:100333446 proto-oncogene serine/threonine-protein kinase P... 31.6 3.2
ath:AT1G71250 GDSL-motif lipase/hydrolase family protein 31.2 3.8
dre:445239 zgc:100927; K06883 30.8 4.2
dre:100003413 Jmjd2al protein-like 30.4 6.9
hsa:6862 T, MGC104817, TFT; T, brachyury homolog (mouse); K101... 29.6 9.3
> tgo:TGME49_106630 tRNA (adenine-N(1)-)-methyltransferase catalytic
subunit, putative (EC:2.1.1.36); K07442 tRNA (adenine-N1-)-methyltransferase
catalytic subunit [EC:2.1.1.36]
Length=360
Score = 87.4 bits (215), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 49/135 (36%), Positives = 69/135 (51%), Gaps = 27/135 (20%)
Query 3 EARIEFDLLGISSHVEAIHRDVCADGFC-------QNVEANTAPA----PI--------- 42
EA +F LG+SS++ HRDVC DGF ++V +N++ P+
Sbjct 211 EAEADFKRLGLSSYLSCYHRDVCTDGFVSVSTASGEDVSSNSSETSRVLPLEEKGKTAGA 270
Query 43 -----APVARAAQQQCGECLPMPHSADGVFLDLPSPWLAIKHASVALKGGGRLVTFSPCL 97
P A ++ G LP S D +FLD+PSPWLA+ H AL+ GGR V FSPC+
Sbjct 271 LADSETPDDGGAAEKVG--LPAAGSIDSLFLDVPSPWLALDHVDAALREGGRFVNFSPCV 328
Query 98 EQLHKTLAAAQEHGF 112
EQ+ + E G+
Sbjct 329 EQVQRVCECLHERGY 343
> cpv:cgd5_2470 GCD14 RNA methylase ; K07442 tRNA (adenine-N1-)-methyltransferase
catalytic subunit [EC:2.1.1.36]
Length=312
Score = 84.7 bits (208), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 45/120 (37%), Positives = 61/120 (50%), Gaps = 23/120 (19%)
Query 7 EFDLLGISSHVEAIHRDVCADGFCQNVEANTAPAPIAPVARAAQQQCGECLPMPHSADGV 66
+F GIS +V HRDVC DGF +P+ + + + G+
Sbjct 183 DFQKYGIS-NVVCQHRDVCKDGFI---------SPLLDITKT-------------TIHGI 219
Query 67 FLDLPSPWLAIKHASVALKGGGRLVTFSPCLEQLHKTLAAAQEHGFQDFQAFEILAKPWG 126
FLDLPSPW+AIKHA L+ G +V FSPC+EQ+ K + + FE+L KPWG
Sbjct 220 FLDLPSPWIAIKHADQVLQKGASIVIFSPCIEQVSKNCNELNLLKYIHIRTFEVLYKPWG 279
Score = 32.7 bits (73), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 9/17 (52%), Positives = 16/17 (94%), Gaps = 0/17 (0%)
Query 222 YQMPMKGHTGYVAYCVR 238
YQ+PM+GHTGY+++ ++
Sbjct 295 YQLPMRGHTGYLSFAIK 311
> xla:379113 trmt61a, MGC53312; tRNA methyltransferase 61 homolog
A; K07442 tRNA (adenine-N1-)-methyltransferase catalytic
subunit [EC:2.1.1.36]
Length=303
Score = 78.6 bits (192), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 41/106 (38%), Positives = 58/106 (54%), Gaps = 2/106 (1%)
Query 45 VARAAQQQCGECLPMPHSADGVFLDLPSPWLAIKHASVALKG-GGRLVTFSPCLEQLHKT 103
V Q C + AD VFLD+PSPW AI HA A+K GGR+ +FSPC+EQ+ +T
Sbjct 157 VTVKNQDVCKHGFGVSDIADAVFLDIPSPWEAIGHAKSAMKTEGGRICSFSPCIEQVQRT 216
Query 104 LAAAQEHGFQDFQAFEILAKPWGA-CISRPTPDQRKHEDDVEEGDV 148
A +EHGF + EIL + + ++ PD + ++ DV
Sbjct 217 CLALEEHGFTEINTLEILLRVYDVRTVNLQVPDLGRAAEEKSSLDV 262
> dre:436593 trmt61a, zgc:86657; tRNA methyltransferase 61 homolog
A (S. cerevisiae) (EC:2.1.1.36); K07442 tRNA (adenine-N1-)-methyltransferase
catalytic subunit [EC:2.1.1.36]
Length=305
Score = 77.4 bits (189), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 40/93 (43%), Positives = 54/93 (58%), Gaps = 2/93 (2%)
Query 45 VARAAQQQCGECLPMPHSADGVFLDLPSPWLAIKHASVALK-GGGRLVTFSPCLEQLHKT 103
V Q C + + AD VFLD+PSPW AI HA ALK GGRL +FSPC+EQ+ +T
Sbjct 157 VTVRNQDVCKDGFGITGVADAVFLDIPSPWEAISHAKKALKYKGGRLCSFSPCIEQVQRT 216
Query 104 LAAAQEHGFQDFQAFEILAKPWGA-CISRPTPD 135
A +HGF++ E+L + ++ P PD
Sbjct 217 CEALLDHGFEEICTLEVLLRVHDVRTVTLPLPD 249
> mmu:328162 Trmt61a, 6720458F09Rik, AI606093, Gcd14, Trm61; tRNA
methyltransferase 61 homolog A (S. cerevisiae) (EC:2.1.1.36);
K07442 tRNA (adenine-N1-)-methyltransferase catalytic
subunit [EC:2.1.1.36]
Length=290
Score = 76.3 bits (186), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 39/89 (43%), Positives = 51/89 (57%), Gaps = 2/89 (2%)
Query 49 AQQQCGECLPMPHSADGVFLDLPSPWLAIKHASVALK-GGGRLVTFSPCLEQLHKTLAAA 107
Q C + H AD VFLD+PSPW A+ HA ALK GGR +FSPC+EQ+ +T A
Sbjct 161 TQDVCCSGFGVVHVADAVFLDIPSPWEAVGHAWDALKVEGGRFCSFSPCIEQVQRTCQAL 220
Query 108 QEHGFQDFQAFEILAKPWGA-CISRPTPD 135
HGF + E+L + + +S P PD
Sbjct 221 AAHGFTELSTLEVLPQVYNVRTVSLPLPD 249
> bbo:BBOV_II005260 18.m06436; hypothetical protein; K07442 tRNA
(adenine-N1-)-methyltransferase catalytic subunit [EC:2.1.1.36]
Length=286
Score = 74.7 bits (182), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 34/65 (52%), Positives = 42/65 (64%), Gaps = 0/65 (0%)
Query 62 SADGVFLDLPSPWLAIKHASVALKGGGRLVTFSPCLEQLHKTLAAAQEHGFQDFQAFEIL 121
S D VF DLPSPW A+++ LK G+LVTFSP +EQ + A E GF + + FEIL
Sbjct 189 SIDAVFFDLPSPWDALENGKQVLKNFGKLVTFSPTVEQSQRMSIALSEAGFIEIKTFEIL 248
Query 122 AKPWG 126
KPWG
Sbjct 249 CKPWG 253
> hsa:115708 TRMT61A, C14orf172, FLJ40452, GCD14, Gcd14p, TRM61,
hTRM61; tRNA methyltransferase 61 homolog A (S. cerevisiae)
(EC:2.1.1.36); K07442 tRNA (adenine-N1-)-methyltransferase
catalytic subunit [EC:2.1.1.36]
Length=289
Score = 73.9 bits (180), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 42/107 (39%), Positives = 54/107 (50%), Gaps = 2/107 (1%)
Query 45 VARAAQQQCGECLPMPHSADGVFLDLPSPWLAIKHASVALK-GGGRLVTFSPCLEQLHKT 103
V Q C + H AD VFLD+PSPW A+ HA ALK GGR +FSPC+EQ+ +T
Sbjct 157 VTVRTQDVCRSGFGVSHVADAVFLDIPSPWEAVGHAWDALKVEGGRFCSFSPCIEQVQRT 216
Query 104 LAAAQEHGFQDFQAFEILAKPWGA-CISRPTPDQRKHEDDVEEGDVS 149
A GF + E+L + + +S P PD D D S
Sbjct 217 CQALAARGFSELSTLEVLPQVYNVRTVSLPPPDLGTGTDGPAGSDTS 263
> tpv:TP02_0831 hypothetical protein
Length=312
Score = 72.4 bits (176), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 36/95 (37%), Positives = 48/95 (50%), Gaps = 3/95 (3%)
Query 61 HSADGVFLDLPSPWLAIKHASVALKGGGRLVTFSPCLEQLHKTLAAAQEHGFQDFQAFEI 120
HS D VFLD+PSPW I + + +K G+LV F P +EQ +K G+ + FE+
Sbjct 193 HSIDSVFLDIPSPWNCIHNVTSVIKHFGKLVIFIPSIEQCNKITECLHSSGYMLIRTFEV 252
Query 121 LAKPWGACISRPTPDQRKHEDDVEEG---DVSHNL 152
L KPWG + ED V G D + NL
Sbjct 253 LTKPWGISFTNSDTADSDDEDPVVTGKQVDNTENL 287
> cel:W02A11.1 hypothetical protein; K07442 tRNA (adenine-N1-)-methyltransferase
catalytic subunit [EC:2.1.1.36]
Length=312
Score = 72.4 bits (176), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 59/192 (30%), Positives = 85/192 (44%), Gaps = 55/192 (28%)
Query 7 EFDLLGISSHVEAIHRDVCADGFCQNVEANTAPAPIAPVARAAQQQCGECLPMPHSADGV 66
EF + G+ S A+ R+VC GF +VE ++ADGV
Sbjct 150 EFKVHGLGSVTTAVVRNVCTSGF--HVE--------------------------NAADGV 181
Query 67 FLDLPSPWLAIKHA--SVALKGGGRLVTFSPCLEQLHKTLAAAQEHGFQDFQAFEILAKP 124
FLD+P+PW AI A S+ GGRLV+FSPCLEQ+ + A GF + E++
Sbjct 182 FLDVPAPWEAIPFAAKSIFRARGGRLVSFSPCLEQVQRACLAMAAAGFVSIETIELV--- 238
Query 125 WGACISRPTPDQRK----HEDDVEE----GDVSHNLKAEKQSERGAQGGQVQQPTPFRDL 176
P+Q+K H +EE GD K+ +R G V+Q +
Sbjct 239 ---------PEQKKIVSQHRQTLEEFEELGDAY-----PKERKRNVDGTIVEQSSSAIPR 284
Query 177 LSHFLQQPSSQP 188
++ + P SQP
Sbjct 285 ITIAIVYPYSQP 296
> pfa:PF13_0087 methyltransferase, putative; K07442 tRNA (adenine-N1-)-methyltransferase
catalytic subunit [EC:2.1.1.36]
Length=316
Score = 70.9 bits (172), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 40/62 (64%), Gaps = 0/62 (0%)
Query 64 DGVFLDLPSPWLAIKHASVALKGGGRLVTFSPCLEQLHKTLAAAQEHGFQDFQAFEILAK 123
D +FLD+P+PWL + + LK G V F PC+EQ++K L A +++ F D + +E++ K
Sbjct 171 DSIFLDMPNPWLCVDNMKKVLKERGVFVIFLPCIEQVYKILEALEKNSFSDIETYELINK 230
Query 124 PW 125
W
Sbjct 231 SW 232
> ath:AT5G14600 tRNA (adenine-N1-)-methyltransferase; K07442 tRNA
(adenine-N1-)-methyltransferase catalytic subunit [EC:2.1.1.36]
Length=318
Score = 65.1 bits (157), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 41/125 (32%), Positives = 61/125 (48%), Gaps = 27/125 (21%)
Query 1 STEARIEFDLLGISSHVEAIHRDVCADGFCQNVEANTAPAPIAPVARAAQQQCGECLPMP 60
+ AR +F+ GISS V RD+ +GF P ++ +
Sbjct 146 AVSAREDFEKTGISSLVTVEVRDIQGEGF---------PEKLSGL--------------- 181
Query 61 HSADGVFLDLPSPWLAIKHASVALKGGGRLVTFSPCLEQLHKTLAAAQEHGFQDFQAFEI 120
AD VFLDLP PWLA+ A+ LK G L +FSPC+EQ+ +T + F + + FE+
Sbjct 182 --ADSVFLDLPQPWLAVPSAAKMLKQDGVLCSFSPCIEQVQRTCEVLRSD-FIEIRTFEV 238
Query 121 LAKPW 125
L + +
Sbjct 239 LLRTY 243
> sce:YJL125C GCD14, TRM61; 1-methyladenosine tRNA methyltransferase
subunit (EC:2.1.1.36); K07442 tRNA (adenine-N1-)-methyltransferase
catalytic subunit [EC:2.1.1.36]
Length=383
Score = 62.4 bits (150), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 42/117 (35%), Positives = 57/117 (48%), Gaps = 17/117 (14%)
Query 13 ISSHVEAIHRDVCADGFCQNVEANTAPAPIAPVARAAQQQCGECLPMPHSADGVFLDLPS 72
I +V HRDVC GF + +T AA +A+ VFLDLP+
Sbjct 158 IDDNVTITHRDVCQGGFLIK-KGDTTSYEFGNNETAASL----------NANVVFLDLPA 206
Query 73 PWLAIKH----ASVALKGGGRLVTFSPCLEQLHKTLAAAQEHGFQDFQAFEILAKPW 125
PW AI H SV K G L FSPC+EQ+ KTL +++G+ D + EI + +
Sbjct 207 PWDAIPHLDSVISVDEKVG--LCCFSPCIEQVDKTLDVLEKYGWTDVEMVEIQGRQY 261
> hsa:9745 ZNF536, KIAA0390; zinc finger protein 536
Length=1300
Score = 33.9 bits (76), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 28/115 (24%), Positives = 45/115 (39%), Gaps = 25/115 (21%)
Query 94 SPCLEQLHKTLAAAQEHGFQDFQAFEILAKPWG---ACISRPTPDQRKHEDDVEEGDVSH 150
S C+E+L AA+ +QA++++A+ +S+ P QR HED + V
Sbjct 500 SSCIERLQAAAKAAEMDPVNSYQAWQLMARGMAMEHGFLSKEHPLQRNHEDTLANAGVLF 559
Query 151 NLKAEKQSERGAQGGQVQQPTPF----------------------RDLLSHFLQQ 183
+ + + GA G + + P RD LSH L Q
Sbjct 560 DKEKREYVLVGADGSKQKMPADLVHSTKVGSQRDLPSKLDPLESSRDFLSHGLNQ 614
> pfa:PF08_0107 VAR; erythrocyte membrane protein 1, PfEMP1; K13850
erythrocyte membrane protein 1
Length=2265
Score = 32.0 bits (71), Expect = 1.9, Method: Composition-based stats.
Identities = 19/46 (41%), Positives = 23/46 (50%), Gaps = 6/46 (13%)
Query 138 KHEDDVEEGDVSHNLKAE----KQSERGAQGGQVQQPTPFRDLLSH 179
KHE D E D ++ E KQ ERG GG+ P+P DL H
Sbjct 793 KHEKD--EADKCKQIQEECNKQKQQERGGPGGRSADPSPPADLDEH 836
> mmu:14812 Grin2b, AW490526, NR2B, Nmdar2b; glutamate receptor,
ionotropic, NMDA2B (epsilon 2); K05210 glutamate receptor,
ionotropic, N-methyl-D-aspartate 2B
Length=1482
Score = 32.0 bits (71), Expect = 2.0, Method: Composition-based stats.
Identities = 22/61 (36%), Positives = 30/61 (49%), Gaps = 1/61 (1%)
Query 35 ANTAPAPIAPVARAAQQQCGECLPMPHSADGVFLDLPSPWLAIKHASVALKGGGRLVTFS 94
A+T P +P AQ++ L HS D F+DL A+ SV+LK GR + S
Sbjct 1276 ASTTKYPQSPTNSKAQKKNRNKLRRQHSYD-TFVDLQKEEAALAPRSVSLKDKGRFMDGS 1334
Query 95 P 95
P
Sbjct 1335 P 1335
> hsa:2904 GRIN2B, MGC142178, MGC142180, NMDAR2B, NR2B, hNR3;
glutamate receptor, ionotropic, N-methyl D-aspartate 2B; K05210
glutamate receptor, ionotropic, N-methyl-D-aspartate 2B
Length=1484
Score = 32.0 bits (71), Expect = 2.0, Method: Composition-based stats.
Identities = 22/61 (36%), Positives = 30/61 (49%), Gaps = 1/61 (1%)
Query 35 ANTAPAPIAPVARAAQQQCGECLPMPHSADGVFLDLPSPWLAIKHASVALKGGGRLVTFS 94
A+T P +P AQ++ L HS D F+DL A+ SV+LK GR + S
Sbjct 1276 ASTTKYPQSPTNSKAQKKNRNKLRRQHSYD-TFVDLQKEEAALAPRSVSLKDKGRFMDGS 1334
Query 95 P 95
P
Sbjct 1335 P 1335
> dre:100333446 proto-oncogene serine/threonine-protein kinase
Pim-1-like
Length=503
Score = 31.6 bits (70), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 35/115 (30%), Positives = 50/115 (43%), Gaps = 18/115 (15%)
Query 130 SRPTPDQRKHEDDVEEG------DVSHNLKAEKQSERGAQGGQVQQPTPFRDLLS---HF 180
S P+PDQ ED ++G D+S K KQ G G V + +D L F
Sbjct 217 SNPSPDQVSQEDSADDGTCLDNNDISCRYKLGKQLGEGG-FGSVYEGIRIQDGLQVAVKF 275
Query 181 LQQ-PSSQPVSKPTTGSSPLQATAA--AAVDVRRKNTLHISSY-----HYQMPMK 227
+Q+ P+ Q VS PL+ T A A+ R N + + + HY M M+
Sbjct 276 VQKTPNMQDVSSSCDQPLPLEITLANMASGGSRCSNIIQLLDWQVFENHYVMVME 330
> ath:AT1G71250 GDSL-motif lipase/hydrolase family protein
Length=374
Score = 31.2 bits (69), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 23/63 (36%), Positives = 32/63 (50%), Gaps = 12/63 (19%)
Query 165 GQVQQPTPFRDLLSHFLQQPSSQPVSKPTT-GSSPLQ----ATAAAAVDVRRKNTLHISS 219
G+ F DLL+ L+ PS P + PTT G+ LQ A+AAA + L +S
Sbjct 79 GRFSNGLTFIDLLARLLEIPSPPPFADPTTSGNRILQGVNYASAAAGI-------LDVSG 131
Query 220 YHY 222
Y+Y
Sbjct 132 YNY 134
> dre:445239 zgc:100927; K06883
Length=349
Score = 30.8 bits (68), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 26/97 (26%), Positives = 40/97 (41%), Gaps = 23/97 (23%)
Query 93 FSPCLEQLHKTLAAAQEHGFQDFQAFEILAKPWGAC--------------------ISRP 132
+ P E+LH+ LA AQ Q + E L K GA +R
Sbjct 243 YRPEYERLHRELAEAQSQKQQ--EQLERLRKDMGAVPMETAAPTEAPDLGGPSDLIFTRG 300
Query 133 TPDQRKHEDDVEEGDVSHNLKAEKQSERGAQGGQVQQ 169
PD+ + E D + D+ H + E+ E GA G +++
Sbjct 301 NPDEAEEEIDSDTDDIDHTV-TEESKEAGAFGNFLKE 336
> dre:100003413 Jmjd2al protein-like
Length=2012
Score = 30.4 bits (67), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 26/90 (28%), Positives = 42/90 (46%), Gaps = 13/90 (14%)
Query 88 GRLVTFSPCLEQLHKTLAAAQEHGFQDFQAFEILAKPWGACI-------SRPTPDQRKHE 140
G+L T C + + K H FQ + K W A S+PTP+ + +
Sbjct 303 GKLATLCSCRKDMVKISMDVFVHKFQPERY-----KLWKAGKDNVPIDHSKPTPEVAEIQ 357
Query 141 DDVEEGDVSHNLKAEKQSERG-AQGGQVQQ 169
+ E+G++S + +AE ERG A+ G Q+
Sbjct 358 KENEKGEISDSERAETLEERGEAEAGDQQK 387
> hsa:6862 T, MGC104817, TFT; T, brachyury homolog (mouse); K10172
brachyury protein
Length=435
Score = 29.6 bits (65), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 16/37 (43%), Positives = 22/37 (59%), Gaps = 2/37 (5%)
Query 173 FRDLLSHFLQQPSSQPVSKPTTGSSPLQATAAAAVDV 209
FR +H+ P + PVS P++ SPL AAAA D+
Sbjct 376 FRGSPAHY--TPLTHPVSAPSSSGSPLYEGAAAATDI 410
Lambda K H
0.316 0.131 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8892779092
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40