bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0452_orf2
Length=169
Score E
Sequences producing significant alignments: (Bits) Value
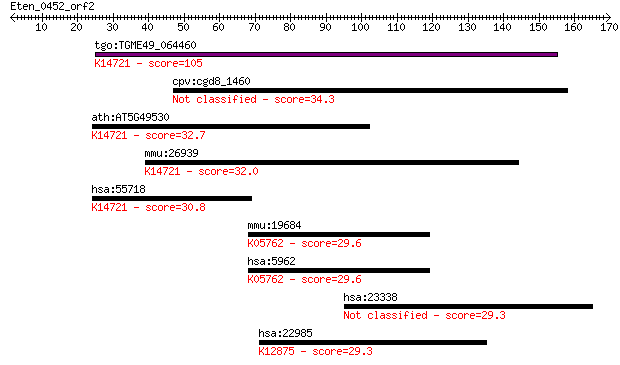
tgo:TGME49_064460 SIN-like domain-containing protein (EC:3.2.1... 105 9e-23
cpv:cgd8_1460 Sex-lethal interacting like 34.3 0.21
ath:AT5G49530 SIN-like family protein; K14721 DNA-directed RNA... 32.7 0.62
mmu:26939 Polr3e, RPC5, Sin; polymerase (RNA) III (DNA directe... 32.0 1.0
hsa:55718 POLR3E, RPC5, SIN; polymerase (RNA) III (DNA directe... 30.8 2.2
mmu:19684 Rdx, AA516625; radixin; K05762 radixin 29.6 4.7
hsa:5962 RDX, DFNB24; radixin; K05762 radixin 29.6 5.5
hsa:23338 PHF15, JADE2, KIAA0239; PHD finger protein 15 29.3
hsa:22985 ACIN1, ACINUS, ACN, DKFZp667N107, KIAA0670, fSAP152;... 29.3 7.2
> tgo:TGME49_064460 SIN-like domain-containing protein (EC:3.2.1.3);
K14721 DNA-directed RNA polymerase III subunit RPC5
Length=836
Score = 105 bits (261), Expect = 9e-23, Method: Compositional matrix adjust.
Identities = 64/169 (37%), Positives = 92/169 (54%), Gaps = 39/169 (23%)
Query 25 HVLKSKVVKGTSCSYAVGLIKEKNLFIIPIHSLLQFKPDFAAFEALQQQQQ-----QQLS 79
HVLKS+ CSYAVG++K+ ++I P+ +LQF+PDF+ + LQQ+ ++LS
Sbjct 233 HVLKSQRALAGDCSYAVGILKKDKIYITPVDHVLQFRPDFSQVDQLQQKASSCPAGRRLS 292
Query 80 SSG----------RRAAHHAD---AAAAAAAEQQQQQ--------------QQQLVQPL- 111
+SG RR D A + AAE Q + Q+ +P
Sbjct 293 ASGDPRASGPGVRRRNEEEKDEKSEAPSGAAEPPQGESAAGGRDPQRPGVPQRSGERPAT 352
Query 112 ------GETRARISHSKQRDIEESEPWLPIDAFYDADSPEAYDLINSLV 154
GE R +ISHS+QRDIEESE W+P++ FYDA++PEA D+I +L
Sbjct 353 VVASSPGEGRGKISHSRQRDIEESEHWIPLETFYDAETPEALDIIRTLC 401
> cpv:cgd8_1460 Sex-lethal interacting like
Length=895
Score = 34.3 bits (77), Expect = 0.21, Method: Composition-based stats.
Identities = 29/130 (22%), Positives = 51/130 (39%), Gaps = 28/130 (21%)
Query 47 KNLFIIPIHSLLQFKPDFAAFEALQQQQQQQLSSSGRRAAHHADAAAAAAAEQQQQQQQQ 106
K L+I PI S+ QF+P+F++ + Q S+ + +A +++ Q
Sbjct 122 KKLYITPISSVQQFRPNFSSID--------QKRSASMNVGMVERLSESADKDERNQSLDD 173
Query 107 LVQPLGETRARISHSKQRDIEES-------------------EPWLPIDAFYDADSPEAY 147
LV G A + H + E W+ I Y ++S E+
Sbjct 174 LVLNHGNN-ATLDHDMNSSSNNNPLYSGNFPLDSGRLYTTGFENWIKIPCIYPSNSYESL 232
Query 148 DLINSLVDNY 157
++I L+D Y
Sbjct 233 EIIKMLMDVY 242
> ath:AT5G49530 SIN-like family protein; K14721 DNA-directed RNA
polymerase III subunit RPC5
Length=689
Score = 32.7 bits (73), Expect = 0.62, Method: Composition-based stats.
Identities = 14/78 (17%), Positives = 38/78 (48%), Gaps = 0/78 (0%)
Query 24 QHVLKSKVVKGTSCSYAVGLIKEKNLFIIPIHSLLQFKPDFAAFEALQQQQQQQLSSSGR 83
+ LK+ + + YAVG++ L + P+H++ Q +P + + ++++Q++ +
Sbjct 191 KQTLKTTWKQPPTLDYAVGVLSGDKLHLNPVHAVAQLRPSMQSLSSDKKKKQEESTEESV 250
Query 84 RAAHHADAAAAAAAEQQQ 101
+ + A+ Q+
Sbjct 251 GTSKKQNKGVQQASTDQK 268
> mmu:26939 Polr3e, RPC5, Sin; polymerase (RNA) III (DNA directed)
polypeptide E; K14721 DNA-directed RNA polymerase III subunit
RPC5
Length=684
Score = 32.0 bits (71), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 27/115 (23%), Positives = 51/115 (44%), Gaps = 22/115 (19%)
Query 39 YAVGLIKEKNLFIIPIHSLLQFKPDFAAFEALQQQQQQQLSSSGRRAAHHA-DAAAAAAA 97
YA L ++ L + P+H +LQ +P F+ + + ++ R AA+ A D++ A
Sbjct 88 YAAALYRQGELHLTPLHGILQLRPSFSYLDKADAKHRE------REAANEAGDSSQDEAE 141
Query 98 EQQQQQQQQLVQPLGETRARISHSKQRDIEE---------SEPWLPIDAFYDADS 143
E +Q + +P E ++QR ++ EPW+ + + DS
Sbjct 142 EDVKQITVRFSRPESE------QARQRRVQSYEFLQKKHAEEPWVHLHYYGMRDS 190
> hsa:55718 POLR3E, RPC5, SIN; polymerase (RNA) III (DNA directed)
polypeptide E (80kD); K14721 DNA-directed RNA polymerase
III subunit RPC5
Length=708
Score = 30.8 bits (68), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 23/45 (51%), Gaps = 1/45 (2%)
Query 24 QHVLKSKVVKGTSCSYAVGLIKEKNLFIIPIHSLLQFKPDFAAFE 68
Q S+ TS YA L ++ L + P+H +LQ +P F+ +
Sbjct 100 QTFCSSQTTSNTS-RYAAALYRQGELHLTPLHGILQLRPSFSYLD 143
> mmu:19684 Rdx, AA516625; radixin; K05762 radixin
Length=583
Score = 29.6 bits (65), Expect = 4.7, Method: Composition-based stats.
Identities = 20/58 (34%), Positives = 30/58 (51%), Gaps = 7/58 (12%)
Query 68 EALQQQQQQQLSSSG-------RRAAHHADAAAAAAAEQQQQQQQQLVQPLGETRARI 118
+AL+ +Q++Q + RRAA A +A A A Q + Q+QL L E A+I
Sbjct 371 KALELEQERQRAKEEAERLDRERRAAEEAKSAIAKQAADQMKNQEQLAAELAEFTAKI 428
> hsa:5962 RDX, DFNB24; radixin; K05762 radixin
Length=583
Score = 29.6 bits (65), Expect = 5.5, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 26/51 (50%), Gaps = 0/51 (0%)
Query 68 EALQQQQQQQLSSSGRRAAHHADAAAAAAAEQQQQQQQQLVQPLGETRARI 118
E + +++ + RRAA A +A A A Q + Q+QL L E A+I
Sbjct 378 ERKRAKEEAERLEKERRAAEEAKSAIAKQAADQMKNQEQLAAELAEFTAKI 428
> hsa:23338 PHF15, JADE2, KIAA0239; PHD finger protein 15
Length=790
Score = 29.3 bits (64), Expect = 5.9, Method: Composition-based stats.
Identities = 24/72 (33%), Positives = 35/72 (48%), Gaps = 2/72 (2%)
Query 95 AAAEQQQQQQQQLVQPLGETRARISHSKQRDIEESEPWLPIDAFYDADSPEAY--DLINS 152
A AE + +++ PL A+ S S E S+P P + YD D +AY +LINS
Sbjct 93 AGAEAIPEPVVRILPPLEGPPAQASPSSTMLGEGSQPDWPGGSRYDLDEIDAYWLELINS 152
Query 153 LVDNYEAPAADD 164
+ E P D+
Sbjct 153 ELKEMERPELDE 164
> hsa:22985 ACIN1, ACINUS, ACN, DKFZp667N107, KIAA0670, fSAP152;
apoptotic chromatin condensation inducer 1; K12875 apoptotic
chromatin condensation inducer in the nucleus
Length=1328
Score = 29.3 bits (64), Expect = 7.2, Method: Composition-based stats.
Identities = 17/64 (26%), Positives = 28/64 (43%), Gaps = 0/64 (0%)
Query 71 QQQQQQQLSSSGRRAAHHADAAAAAAAEQQQQQQQQLVQPLGETRARISHSKQRDIEESE 130
+ Q+Q+ L GR +A + A QQQ+++ + PL E I S+ +
Sbjct 327 RSQEQEVLERGGRFTRSQEEARKSHLARQQQEKEMKTTSPLEEEEREIKSSQGLKEKSKS 386
Query 131 PWLP 134
P P
Sbjct 387 PSPP 390
Lambda K H
0.314 0.126 0.350
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4222647260
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40