bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0459_orf1
Length=85
Score E
Sequences producing significant alignments: (Bits) Value
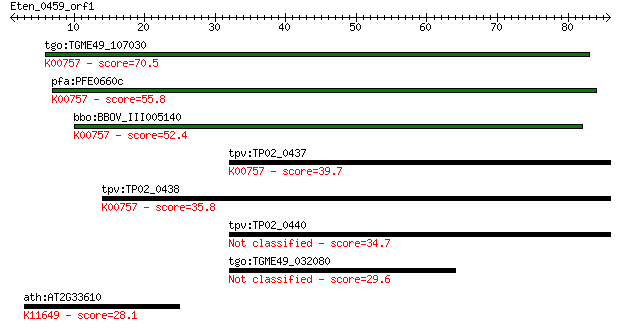
tgo:TGME49_107030 purine nucleoside phosphorylase (EC:2.4.2.3)... 70.5 1e-12
pfa:PFE0660c purine nucleotide phosphorylase, putative (EC:2.4... 55.8 3e-08
bbo:BBOV_III005140 17.m07459; phosphorylase family protein (EC... 52.4 3e-07
tpv:TP02_0437 purine nucleoside phosphorylase; K00757 uridine ... 39.7 0.003
tpv:TP02_0438 purine nucleoside phosphorylase; K00757 uridine ... 35.8 0.031
tpv:TP02_0440 purine nucleoside phosphorylase 34.7 0.075
tgo:TGME49_032080 hypothetical protein 29.6 2.7
ath:AT2G33610 ATSWI3B (SWITCH SUBUNIT 3); DNA binding; K11649 ... 28.1 6.7
> tgo:TGME49_107030 purine nucleoside phosphorylase (EC:2.4.2.3);
K00757 uridine phosphorylase [EC:2.4.2.3]
Length=247
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 32/77 (41%), Positives = 44/77 (57%), Gaps = 0/77 (0%)
Query 6 NLTNWSKFTDIIDCEFQSLXLVGIGRGIETGGIATVDGSPLQWDQANYDPSGNTVAEGKK 65
L +SK D+ID E + + RGI T GI VDGSPLQWD+ +YD +G GK+
Sbjct 171 KLEMYSKCCDVIDMEMSGVLGLCQARGIATCGILAVDGSPLQWDEGDYDATGVKATTGKE 230
Query 66 KMLTVAIHTCTKLTREY 82
M+ + + C L R+Y
Sbjct 231 NMIKITLKACANLRRQY 247
> pfa:PFE0660c purine nucleotide phosphorylase, putative (EC:2.4.2.3);
K00757 uridine phosphorylase [EC:2.4.2.3]
Length=245
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 46/78 (58%), Gaps = 3/78 (3%)
Query 7 LTNWSKF-TDIIDCEFQSLXLVGIGRGIETGGIATVDGSPLQWDQANYDPSGNTVAEGKK 65
L ++SK +++ E +L ++G R ++TGGI VDG P +WD+ ++D N V +
Sbjct 170 LEDYSKANAAVVEMELATLMVIGTLRKVKTGGILIVDGCPFKWDEGDFD--NNLVPHQLE 227
Query 66 KMLTVAIHTCTKLTREYS 83
M+ +A+ C KL +Y+
Sbjct 228 NMIKIALGACAKLATKYA 245
> bbo:BBOV_III005140 17.m07459; phosphorylase family protein (EC:2.4.2.3);
K00757 uridine phosphorylase [EC:2.4.2.3]
Length=253
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 22/74 (29%), Positives = 43/74 (58%), Gaps = 2/74 (2%)
Query 10 WSKFTDII--DCEFQSLXLVGIGRGIETGGIATVDGSPLQWDQANYDPSGNTVAEGKKKM 67
++K+ +++ D + ++ L+ G+ G + T+DG PL+WD+ NY P + GK M
Sbjct 177 YAKYKEVVIEDTDNSAVLLLCNLYGVWGGALCTIDGCPLEWDKGNYQPHTEKMQHGKTNM 236
Query 68 LTVAIHTCTKLTRE 81
+ +A+H L+R+
Sbjct 237 VKLAMHAAANLSRQ 250
> tpv:TP02_0437 purine nucleoside phosphorylase; K00757 uridine
phosphorylase [EC:2.4.2.3]
Length=256
Score = 39.7 bits (91), Expect = 0.003, Method: Composition-based stats.
Identities = 18/54 (33%), Positives = 27/54 (50%), Gaps = 0/54 (0%)
Query 32 GIETGGIATVDGSPLQWDQANYDPSGNTVAEGKKKMLTVAIHTCTKLTREYSGN 85
G + G + T+DG+PL W Y V G +KM +A +KL+ +Y N
Sbjct 201 GAKAGAVMTIDGTPLYWSSGGYIVGEKIVDVGIEKMTLLASKAASKLSNKYRNN 254
> tpv:TP02_0438 purine nucleoside phosphorylase; K00757 uridine
phosphorylase [EC:2.4.2.3]
Length=254
Score = 35.8 bits (81), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 17/72 (23%), Positives = 34/72 (47%), Gaps = 0/72 (0%)
Query 14 TDIIDCEFQSLXLVGIGRGIETGGIATVDGSPLQWDQANYDPSGNTVAEGKKKMLTVAIH 73
TD + E ++ ++ G + G I T+DG PL W + V +G + M+ + +
Sbjct 183 TDTDEAETATIFVLSSLYGAKAGAIMTIDGFPLLWSSGKSNSGKKEVDKGIENMMRIGLK 242
Query 74 TCTKLTREYSGN 85
+ L++ + N
Sbjct 243 AASTLSKRFFNN 254
> tpv:TP02_0440 purine nucleoside phosphorylase
Length=254
Score = 34.7 bits (78), Expect = 0.075, Method: Compositional matrix adjust.
Identities = 16/54 (29%), Positives = 26/54 (48%), Gaps = 0/54 (0%)
Query 32 GIETGGIATVDGSPLQWDQANYDPSGNTVAEGKKKMLTVAIHTCTKLTREYSGN 85
G + + TVDG PL W + Y V G +KM+ + + + LT+ + N
Sbjct 201 GAKAASVLTVDGFPLLWARGGYLVGEKAVDIGIEKMMGLGVKAASTLTKRFFNN 254
> tgo:TGME49_032080 hypothetical protein
Length=10329
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 16/32 (50%), Gaps = 0/32 (0%)
Query 32 GIETGGIATVDGSPLQWDQANYDPSGNTVAEG 63
G+ G ++T G PL W PSG T A G
Sbjct 6506 GVAGGSVSTFYGPPLGWTDGALAPSGWTAATG 6537
> ath:AT2G33610 ATSWI3B (SWITCH SUBUNIT 3); DNA binding; K11649
SWI/SNF related-matrix-associated actin-dependent regulator
of chromatin subfamily C
Length=469
Score = 28.1 bits (61), Expect = 6.7, Method: Composition-based stats.
Identities = 10/22 (45%), Positives = 16/22 (72%), Gaps = 0/22 (0%)
Query 3 HIENLTNWSKFTDIIDCEFQSL 24
H+ + ++W +TDI DCE +SL
Sbjct 49 HVPSYSSWFSWTDINDCEVRSL 70
Lambda K H
0.314 0.133 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2035463248
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40