bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0516_orf6
Length=623
Score E
Sequences producing significant alignments: (Bits) Value
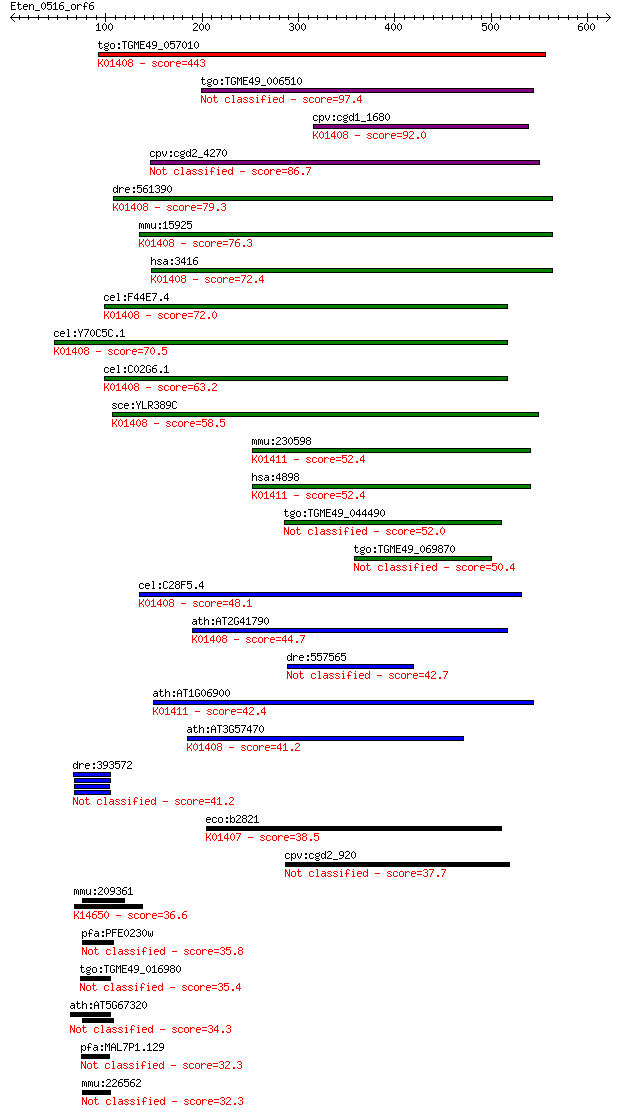
tgo:TGME49_057010 insulysin, putative (EC:3.4.24.56); K01408 i... 443 9e-124
tgo:TGME49_006510 peptidase M16 domain containing protein (EC:... 97.4 2e-19
cpv:cgd1_1680 insulinase like protease, signal peptide ; K0140... 92.0 7e-18
cpv:cgd2_4270 secreted insulinase-like peptidase 86.7 2e-16
dre:561390 ide, MGC162603, zgc:162603; insulin-degrading enzym... 79.3 4e-14
mmu:15925 Ide, 1300012G03Rik, 4833415K22Rik, AA675336, AI50753... 76.3 3e-13
hsa:3416 IDE, FLJ35968, INSULYSIN; insulin-degrading enzyme (E... 72.4 5e-12
cel:F44E7.4 hypothetical protein; K01408 insulysin [EC:3.4.24.56] 72.0 6e-12
cel:Y70C5C.1 hypothetical protein; K01408 insulysin [EC:3.4.24... 70.5 2e-11
cel:C02G6.1 hypothetical protein; K01408 insulysin [EC:3.4.24.56] 63.2 3e-09
sce:YLR389C STE23; Metalloprotease involved, with homolog Axl1... 58.5 7e-08
mmu:230598 Nrd1, 2600011I06Rik, AI875733, MGC25477, NRD-C; nar... 52.4 6e-06
hsa:4898 NRD1, hNRD1, hNRD2; nardilysin (N-arginine dibasic co... 52.4 6e-06
tgo:TGME49_044490 insulin-degrading enzyme, putative (EC:3.4.2... 52.0 7e-06
tgo:TGME49_069870 hypothetical protein 50.4 2e-05
cel:C28F5.4 hypothetical protein; K01408 insulysin [EC:3.4.24.56] 48.1 1e-04
ath:AT2G41790 peptidase M16 family protein / insulinase family... 44.7 0.001
dre:557565 nrd1, si:dkey-171o17.4; nardilysin (N-arginine diba... 42.7 0.004
ath:AT1G06900 catalytic/ metal ion binding / metalloendopeptid... 42.4 0.006
ath:AT3G57470 peptidase M16 family protein / insulinase family... 41.2 0.011
dre:393572 traf3ip1, Elipsa, MGC63522, zgc:63522; TNF receptor... 41.2 0.014
eco:b2821 ptrA, ECK2817, JW2789, ptr; protease III (EC:3.4.24.... 38.5 0.072
cpv:cgd2_920 peptidase'insulinase-like peptidase' 37.7 0.14
mmu:209361 Taf3, 140kDa, 4933439M23Rik, AW539625, TAF140, TAFI... 36.6 0.31
pfa:PFE0230w conserved Plasmodium protein, unknown function 35.8 0.56
tgo:TGME49_016980 hypothetical protein 35.4 0.62
ath:AT5G67320 HOS15; HOS15 (high expression of osmotically res... 34.3 1.7
pfa:MAL7P1.129 conserved Plasmodium protein, unknown function 32.3 5.5
mmu:226562 Prrc2c, 1810043M20Rik, 9630039I18Rik, A630006J20, B... 32.3 6.5
> tgo:TGME49_057010 insulysin, putative (EC:3.4.24.56); K01408
insulysin [EC:3.4.24.56]
Length=953
Score = 443 bits (1140), Expect = 9e-124, Method: Compositional matrix adjust.
Identities = 213/466 (45%), Positives = 315/466 (67%), Gaps = 3/466 (0%)
Query 92 RERERERERERERPNAFRMPPPLLHIPKASELEILPGLLGLNEPELISEQGGNAGTAVWW 151
+E+ R E P A+++PP L H+P+ +L +LP L G++ PEL+ + + G AVWW
Sbjct 485 KEQRRRLETVTPSPGAYKIPPALKHVPRPEDLHLLPALGGMSIPELLGDSNTSGGHAVWW 544
Query 152 QGQGFSALPRVAVQLSGSIAKDKADLLSRTQGSVALAAIAEHLQEETADFQYCGITHSLA 211
QGQG +PRV + + + ++ SRTQ ++ +AA+AE L EET D + CGI+HS+
Sbjct 545 QGQGTLPVPRVHANIKARTQRSRTNMASRTQATLLMAALAEQLDEETVDLKQCGISHSVG 604
Query 212 FKGTGFHMTFEGYTKTQLDKLMSHVASVLRDPSVVESERFERVKQKQIKLLADPATNMAF 271
G G + F YT QL ++M+ VAS ++DP V E +RF+R+KQ+ I+ L D A+ +A+
Sbjct 605 VSGDGLLLAFAAYTPKQLRQVMAVVASKIQDPQV-EQDRFDRIKQRMIEELEDSASQVAY 663
Query 272 EHALQAAAILTRNDAFSRMDLLNALEQ--TTYDDSIAKLSELKNVHVDAFVMGNIDRDQS 329
EHA+ AA++L RNDA SR DLL L+ T+ ++++ +LK VH DAF+MGNID+ +
Sbjct 664 EHAIAAASVLLRNDANSRKDLLRLLKSSSTSLNETLKTFRDLKAVHADAFIMGNIDKADA 723
Query 330 LLLTESFLEQAGFTPIEHDDAVESLAMEQKQTIEATLANPIKGDKDHASLVQFQLGVPSI 389
+ +SFL+ +GFT I DA +SL ++Q+ IEA +ANPI D +HA++VQ+QLGVPSI
Sbjct 724 NSVVQSFLQDSGFTQIPMKDAAQSLVVDQRAPIEALIANPIPKDVNHATVVQYQLGVPSI 783
Query 390 EERVNLAILSQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGSKSHPDEVVK 449
EERVNLA+L Q LNRR++D LRTE QLGYI GA+ +S L+C +EGS+ HPDE+
Sbjct 784 EERVNLAVLGQMLNRRLFDRLRTEEQLGYIVGARSYIDSSVESLRCVLEGSRKHPDEIAD 843
Query 450 MIDAELAKAKDYISNMPEAELSRWKEAAHAKLTKVEANFSEDFKKSADEIFSHANCFSKR 509
+ID EL K D++ ++ + EL WKE+A A+L K F E+F +S +I +H +CF+KR
Sbjct 844 LIDKELWKMNDHLQSISDGELDHWKESARAELEKPTETFYEEFGRSWGQIANHGHCFNKR 903
Query 510 DLEVKYLDNDFSRKHLLRTFTKLSDPSRRMVVKLIADLEPAKEVTL 555
DLE+ YL+ +F+RK L RT+TKL +P+RR+V ++ + P +L
Sbjct 904 DLELIYLNTEFNRKQLSRTYTKLLEPTRRLVGFVLNHVTPCLNKSL 949
> tgo:TGME49_006510 peptidase M16 domain containing protein (EC:4.1.1.70
3.4.24.13 3.4.24.56 3.4.21.10 3.4.24.35 3.2.1.91)
Length=2435
Score = 97.4 bits (241), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 88/356 (24%), Positives = 170/356 (47%), Gaps = 13/356 (3%)
Query 199 ADFQYCGITHSLAFKGTGFHMTFEGYTKTQLDKLMSHVASVLRDPS-VVESERFERVKQK 257
A FQ CG+ ++F + + +++ +++ + VL++ V+ F ++
Sbjct 1011 ASFQGCGVDLLMSFTNGALVLEIQAFSEL-FAPVLARLIEVLKESQDNVKQSDFNKIFNT 1069
Query 258 QIKLLADPATNMAFEHALQAAAILTRNDAFSRMDLLNALEQTT--YDDSIAKLSEL--KN 313
L+D +T FE AL A + R + FS++DL +A+ + ++D L ++ KN
Sbjct 1070 LKVQLSDFSTVTPFELALDVALSVVRRNRFSQLDLRSAVTDASSQFEDFKVFLEKVLTKN 1129
Query 314 VHVDAFVMGNIDRDQSLLLTESFLEQAGFTPIEHDDAVESLAMEQKQTIEATLANPIKGD 373
+D F+MG+ID +++ L E F P+ ++ S + IE +NPI D
Sbjct 1130 A-LDVFIMGDIDYEEARKLAEDFRAALSKQPLPFSESAGSEILNLADDIEIRFSNPIPED 1188
Query 374 KDHASLVQFQL-GVPSIEERVNLAILSQFLNRRIYDSLRTEAQLGYIAGAKESQAASTAL 432
+A + + P + E V +++ + ++ +D++RT GY+A A +
Sbjct 1189 ATNAYVSLYVTHPPPDMMEMVVYSLIGEVISSPFFDTIRTHWMDGYVAAAAVREVPPAMT 1248
Query 433 LQCFVEGSKSHPDEVVKMIDAELAKAKDYI--SNMPEAELSRWKEAAHAKLTKVEANFSE 490
L V+GS+ PDE+ + + A LA+ ++ I S EA L R + + +K + +FS+
Sbjct 1249 LATIVQGSQRKPDELERHVCAFLAEMEENIGSSMTTEAFLERLRWLSSSKFHRSATSFSD 1308
Query 491 DFKKSADEIFSHANCFSKRDLEVKYLDNDFSRKHLLRTF-TKLSDPS--RRMVVKL 543
F + +I S CF + L + S +L+++ L D + +R+ VK+
Sbjct 1309 YFGEVTSQIASRNFCFIREQLARLATEKFLSCPAILKSYMNSLVDRANRKRITVKI 1364
> cpv:cgd1_1680 insulinase like protease, signal peptide ; K01408
insulysin [EC:3.4.24.56]
Length=1033
Score = 92.0 bits (227), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 64/229 (27%), Positives = 111/229 (48%), Gaps = 8/229 (3%)
Query 316 VDAFVMGNIDRDQSLLLTESFLEQAGFTPIEHDDAVESLAMEQKQTIEATLANPIKGDKD 375
+ A++ GNI +++SL L E F+ + + +++ + + I+ L NP+ D +
Sbjct 761 IIAYLQGNISKNKSLYLIEKFVLNSKILSLNDKYSMKKKIHKLTRPIDIALINPVFEDIN 820
Query 376 HASLVQFQLGVPSIEERVNLAILSQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQC 435
+ L +Q GVPS EE+++L L + IYD+LRT QLGYI A +ST LL
Sbjct 821 NTVLAFYQFGVPSFEEKLHLMALQPIIQGYIYDNLRTNKQLGYIIFANIVPISSTRLLVV 880
Query 436 FVEGSKSHPDE----VVKMIDAELAKAKDYISNMPEAELSRWKEAAHAKLTKVEANFSED 491
VEG ++ E +++ E + K + NM K A + + +F++
Sbjct 881 GVEGDNNNSVEKIESIIRNTLYEFSTRK--LGNMESHMFEDIKSALIQEAKSIGNSFNQK 938
Query 492 FKKSADEIFSHANCFSKRDLE--VKYLDNDFSRKHLLRTFTKLSDPSRR 538
DEI + +L+ + Y++N + +HL TF+KL + R
Sbjct 939 LNHYWDEIRYVGDLSESFNLQRAIDYINNKMTIEHLYNTFSKLINSKER 987
> cpv:cgd2_4270 secreted insulinase-like peptidase
Length=1257
Score = 86.7 bits (213), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 102/429 (23%), Positives = 193/429 (44%), Gaps = 36/429 (8%)
Query 146 GTAVWWQGQGFSALPRVAVQLSGSIAKDKADLLSRTQGSVALAAIAEHLQEETAD----- 200
G ++W+G +P + + L + D+ + + S+ +A + LQ D
Sbjct 692 GLRIFWKGP-IHTVPTINLTLVQRLPN--KDVSNNVRVSL-IANLHAQLQNSKMDYILSS 747
Query 201 FQYCGITHSLAFKGTGFHMTFEGYTKT---QLDKLMSHVASVLRDPSVVESERFERVKQK 257
F+ CG+ +++ F + + Y+ ++KL +++ S R P+ E E +
Sbjct 748 FKLCGLEADISYSRGRFVINVQSYSSNFEDIIEKLSNYLVSESRLPTKTEFETALTNLKS 807
Query 258 QIKLLADPATNMAFEHALQAAAILTRNDAFSRMDLLNALEQT--TYDDSIAKLSELKNV- 314
+I L+D MA++ A A + ++ +SR+ L +L++T T+D+ I K+ ++ +V
Sbjct 808 EILNLSDL---MAYDVATDVAQSVYLSNYYSRLQLRESLQKTEITFDEYIEKIKDIFSVG 864
Query 315 HVDAFVMGNIDRDQSLLLTESFLEQAGFTPIEHDDAVESLAMEQKQTIEATLANPIKGDK 374
+ DA ++GNI ++S+ L + I + +A+ + I NPI DK
Sbjct 865 YFDALIVGNIGYEKSIKLVSRMVGSLVTKKIPYSNAIHDGILNVSGDIHIKANNPISSDK 924
Query 375 DHASLVQFQLGVPSIEERVNLAILS---QFLNRRIYDSLRTEAQLGYIAGA-KESQAAST 430
++A + F P + + ++++I S + LN YD+LRTE Q GY+A A + +
Sbjct 925 NNAVVAHFL--TPPV-DLIDVSIYSSIGEILNSPFYDTLRTEWQDGYVAFATTKYETPII 981
Query 431 ALLQCFVEGSKSHPDEVVKMIDAELAKAKDYISNMPEAELS------RWKEAAHAKLTKV 484
+L+ K V M A +KD ++ E S RW + K+
Sbjct 982 SLIGAVQSAEKLSETLVCHMFSALKKVSKDVEEDLKEISKSEFEDKIRWFGLSKYSSQKL 1041
Query 485 EANFSEDFKKSADEIFSHANCFSKRDLEVKYLDNDFSRKHL-LRTFTKLSDPS---RRMV 540
++ F++ + + SH CF K L S ++ + KL PS R ++
Sbjct 1042 DS-FTKYIEHFGKLVVSHELCFEKNKLIENATQAFISEPNIYIEKLNKLIKPSSSRRLVI 1100
Query 541 VKLIADLEP 549
V+LI + P
Sbjct 1101 VELIGNKSP 1109
> dre:561390 ide, MGC162603, zgc:162603; insulin-degrading enzyme
(EC:3.4.24.56); K01408 insulysin [EC:3.4.24.56]
Length=978
Score = 79.3 bits (194), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 91/466 (19%), Positives = 194/466 (41%), Gaps = 21/466 (4%)
Query 108 FRMPPPLLHIPKASELEILPGLLGLNEPELISEQGGNAGTAVWWQGQGFSALPRVAVQLS 167
F++P IP + EI P L + P + A + VW++ LP+ +
Sbjct 481 FKLPMKNEFIP--TNFEIYP--LEKDSPSAPTLIKDTAMSKVWFKQDDKFFLPKACLNFE 536
Query 168 GSIAKDKADLLSRTQGSVALAAIAEHLQEETADFQYCGITHSLAFKGTGFHMTFEGYTKT 227
D L + L + + L E + G+++ L G +++ +GY
Sbjct 537 FFSPFAYVDPLHCNMAYLYLELLKDSLNEYAYAAELAGLSYDLQNTVYGMYLSVKGYNDK 596
Query 228 Q---LDKLMSHVASVLRDPSVVESERFERVKQKQIKLLADPATNMAFEHALQAAAILTRN 284
Q L K++ +A+ ++ +RF+ +K+ ++ L + +HA+ +L
Sbjct 597 QHILLKKIIEKMATF-----EIDEKRFDIIKEAYMRSLNNFRAEQPHQHAMYYLRLLMTE 651
Query 285 DAFSRMDLLNALEQTTYDDSIAKLSEL-KNVHVDAFVMGNIDRDQSL----LLTESFLEQ 339
A+++ +L +AL+ T A + +L +H++A + GNI + +L +L ++ +E
Sbjct 652 VAWTKDELRDALDDVTLPRLKAFIPQLLSRLHIEALLHGNITKQSALEMMQMLEDTLIEH 711
Query 340 AGFTPIEHDDAVESLAMEQKQTIEATLANPIKGDKDHASLVQFQLGVPSIEERVNLAILS 399
A P+ + ++ + + + +Q + + E + L +
Sbjct 712 AHTKPLLPSQLIRYREVQVPDGGWYVYQQRNEVHNNCGIEIYYQTDMQNTHENMLLELFC 771
Query 400 QFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGSKSHPDEVVKMIDAELAKAK 459
Q ++ +++LRT+ QLGYI + +A L+ ++ K+ P + ++A L +
Sbjct 772 QIISEPCFNTLRTKEQLGYIVFSGPRRANGVQGLRFIIQSEKA-PHYLESRVEAFLKTME 830
Query 460 DYISNMPEAELSRWKEAAHAKLTKVEANFSEDFKKSADEIFSHANCFSKRDLEVKYLDND 519
+ M + + +A + + + K EI S F + ++EV YL
Sbjct 831 KSVEEMGDEAFQKHIQALAIRRLDKPKKLAAECAKYWGEIISQQYNFDRDNIEVAYLKT- 889
Query 520 FSRKHLLRTFTKL--SDPSRRMVVKLIADLEPAKEVTLIGEAKGQD 563
+++H+++ + L D RR V + L+GE Q+
Sbjct 890 LTKEHIMQFYRDLLAIDAPRRHKVSVHVLSREMDSCPLVGEFPAQN 935
> mmu:15925 Ide, 1300012G03Rik, 4833415K22Rik, AA675336, AI507533;
insulin degrading enzyme (EC:3.4.24.56); K01408 insulysin
[EC:3.4.24.56]
Length=1019
Score = 76.3 bits (186), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 86/439 (19%), Positives = 181/439 (41%), Gaps = 20/439 (4%)
Query 135 PELISEQGGNAGTAVWWQGQGFSALPRVAVQLSGSIAKDKADLLSRTQGSVALAAIAEHL 194
P LI + A + +W++ LP+ + D L + L + + L
Sbjct 548 PALIKD---TAMSKLWFKQDDKFFLPKACLNFEFFSPFAYVDPLHCNMAYLYLELLKDSL 604
Query 195 QEETADFQYCGITHSLAFKGTGFHMTFEGYTKTQ---LDKLMSHVASVLRDPSVVESERF 251
E + G+++ L G +++ +GY Q L K+ +A+ ++ +RF
Sbjct 605 NEYAYAAELAGLSYDLQNTIYGMYLSVKGYNDKQPILLKKITEKMATF-----EIDKKRF 659
Query 252 ERVKQKQIKLLADPATNMAFEHALQAAAILTRNDAFSRMDLLNALEQTTYDDSIAKLSEL 311
E +K+ ++ L + +HA+ +L A+++ +L AL+ T A + +L
Sbjct 660 EIIKEAYMRSLNNFRAEQPHQHAMYYLRLLMTEVAWTKDELKEALDDVTLPRLKAFIPQL 719
Query 312 -KNVHVDAFVMGNIDRDQSL----LLTESFLEQAGFTPIEHDDAVESLAMEQKQTIEATL 366
+H++A + GNI + +L ++ ++ +E A P+ V ++
Sbjct 720 LSRLHIEALLHGNITKQAALGVMQMVEDTLIEHAHTKPLLPSQLVRYREVQLPDRGWFVY 779
Query 367 ANPIKGDKDHASLVQFQLGVPSIEERVNLAILSQFLNRRIYDSLRTEAQLGYIAGAKESQ 426
+ + + +Q + S E + L + Q ++ +++LRT+ QLGYI + +
Sbjct 780 QQRNEVHNNCGIEIYYQTDMQSTSENMFLELFCQIISEPCFNTLRTKEQLGYIVFSGPRR 839
Query 427 AASTALLQCFVEGSKSHPDEVVKMIDAELAKAKDYISNMPEAELSRWKEAAHAKLTKVEA 486
A L+ ++ K P + ++A L + I +M E + +A +
Sbjct 840 ANGIQGLRFIIQSEKP-PHYLESRVEAFLITMEKAIEDMTEEAFQKHIQALAIRRLDKPK 898
Query 487 NFSEDFKKSADEIFSHANCFSKRDLEVKYLDNDFSRKHLLRTFTKL--SDPSRRMVVKLI 544
S + K EI S + + ++EV YL ++ ++R + ++ D RR V +
Sbjct 899 KLSAECAKYWGEIISQQYNYDRDNIEVAYLKT-LTKDDIIRFYQEMLAVDAPRRHKVSVH 957
Query 545 ADLEPAKEVTLIGEAKGQD 563
++GE Q+
Sbjct 958 VLAREMDSCPVVGEFPSQN 976
> hsa:3416 IDE, FLJ35968, INSULYSIN; insulin-degrading enzyme
(EC:3.4.24.56); K01408 insulysin [EC:3.4.24.56]
Length=464
Score = 72.4 bits (176), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 89/433 (20%), Positives = 182/433 (42%), Gaps = 29/433 (6%)
Query 147 TAVWWQGQGFSALPRVAVQLSGSIAKDKADLLSRTQGSVALAAIAEHLQEETADFQYCGI 206
+ +W++ LP+ + D L + L + + L E + G+
Sbjct 2 SKLWFKQDDKFFLPKACLNFEFFSPFAYVDPLHCNMAYLYLELLKDSLNEYAYAAELAGL 61
Query 207 THSLAFKGTGFHMTFEGYTKTQ---LDKLMSHVASVLRDPSVVESERFERVKQKQIKLLA 263
++ L G +++ +GY Q L K++ +A+ ++ +RFE +K+ ++ L
Sbjct 62 SYDLQNTIYGMYLSVKGYNDKQPILLKKIIEKMATF-----EIDEKRFEIIKEAYMRSLN 116
Query 264 DPATNMAFEHALQAAAILTRNDAFSRMDLLNALEQTTYDDSIAKLSEL-KNVHVDAFVMG 322
+ +HA+ +L A+++ +L AL+ T A + +L +H++A + G
Sbjct 117 NFRAEQPHQHAMYYLRLLMTEVAWTKDELKEALDDVTLPRLKAFIPQLLSRLHIEALLHG 176
Query 323 NIDRDQSLLLTESFLEQAGFTPIEHDDAVESLAMEQKQTIEATLANPIKG-------DKD 375
NI + +L ++ T IEH L + + E L P +G ++
Sbjct 177 NITKQAAL----GIMQMVEDTLIEHAHTKPLLPSQLVRYREVQL--PDRGWFVYQQRNEV 230
Query 376 HASL---VQFQLGVPSIEERVNLAILSQFLNRRIYDSLRTEAQLGYIAGAKESQAASTAL 432
H + + +Q + S E + L + Q ++ +++LRT+ QLGYI + +A
Sbjct 231 HNNCGIEIYYQTDMQSTSENMFLELFCQIISEPCFNTLRTKEQLGYIVFSGPRRANGIQG 290
Query 433 LQCFVEGSKSHPDEVVKMIDAELAKAKDYISNMPEAELSRWKEAAHAKLTKVEANFSEDF 492
L+ F+ S+ P + ++A L + I +M E + +A + S +
Sbjct 291 LR-FIIQSEKPPHYLESRVEAFLITMEKSIEDMTEEAFQKHIQALAIRRLDKPKKLSAEC 349
Query 493 KKSADEIFSHANCFSKRDLEVKYLDNDFSRKHLLRTFTKL--SDPSRRMVVKLIADLEPA 550
K EI S F + + EV YL +++ +++ + ++ D RR V +
Sbjct 350 AKYWGEIISQQYNFDRDNTEVAYLKT-LTKEDIIKFYKEMLAVDAPRRHKVSVHVLAREM 408
Query 551 KEVTLIGEAKGQD 563
++GE Q+
Sbjct 409 DSCPVVGEFPCQN 421
> cel:F44E7.4 hypothetical protein; K01408 insulysin [EC:3.4.24.56]
Length=1051
Score = 72.0 bits (175), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 92/430 (21%), Positives = 177/430 (41%), Gaps = 18/430 (4%)
Query 99 ERERERPNAFRMPPPLLHIPK-----ASELEILPGLLGLNE-PELISEQGGNAGTAVWWQ 152
E ++ NA + LH+P+ A+ + P NE P LIS+ G + VW++
Sbjct 531 ETMKKYENALKTSHHALHLPEKNEYIATNFDQKPRESVKNEHPRLISDDGW---SRVWFK 587
Query 153 GQGFSALPRVAVQLSGSIAKDKADLLSRTQGSVALAAIAEHLQEETADFQYCGITHSLAF 212
+P+ +L+ + + S+ L +++ L EET + G+ L
Sbjct 588 QDDEYNMPKQETKLALTTPMVAQNPRMSLLSSLWLWCLSDTLAEETYNADLAGLKCQLES 647
Query 213 KGTGFHMTFEGYTKTQLDKLMSHVASVLRDPSVVESERFERVKQKQIKLLADPATNMAFE 272
G M GY + Q H+A+ + + + + RF+ + + + L + A + +
Sbjct 648 SPFGVQMRVYGYDEKQA-LFAKHLANRMTNFKI-DKTRFDVLFESLKRALTNHAFSQPYL 705
Query 273 HALQAAAILTRNDAFSRMDLLNALEQTTYDDSIAKLSE-LKNVHVDAFVMGNIDRDQSLL 331
+L + +S+ LL + T +D E L+ H++ FV GN +++
Sbjct 706 LTQHYNQLLIVDKVWSKEQLLAVCDSVTLEDVQGFAKEMLQAFHMELFVHGNSTEKEAIQ 765
Query 332 LTESFLE-----QAGFTPIEHDDAVESLAMEQKQTIEATLANPIKGDKDHASLVQFQLGV 386
L++ ++ P+ ++ ++ E + K V +Q+GV
Sbjct 766 LSKELMDVLKSAAPNSRPLYRNEHNPRRELQLNNGDEYVYRHLQKTHDVGCVEVTYQIGV 825
Query 387 PSIEERVNLAILSQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGSKSHPDE 446
+ + + ++ Q + +++LRT LGYI T L V+G KS D
Sbjct 826 QNTYDNAVVGLIDQLIREPAFNTLRTNEALGYIVWTGSRLNCGTVALNVIVQGPKS-VDH 884
Query 447 VVKMIDAELAKAKDYISNMPEAELSRWKEAAHAKLTKVEANFSEDFKKSADEIFSHANCF 506
V++ I+ L + I+ MP+ E A+L + S F++ +EI F
Sbjct 885 VLERIEVFLESVRKEIAEMPQEEFDNQVSGMIARLEEKPKTLSSRFRRFWNEIECRQYNF 944
Query 507 SKRDLEVKYL 516
++R+ EV L
Sbjct 945 ARREEEVALL 954
> cel:Y70C5C.1 hypothetical protein; K01408 insulysin [EC:3.4.24.56]
Length=985
Score = 70.5 bits (171), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 100/483 (20%), Positives = 204/483 (42%), Gaps = 31/483 (6%)
Query 47 LAVLVPSVARNPHLFLLLSTRRRNKTKNSRERERERERERERERERERERERERERERPN 106
L++L+PS + ++S + + + N+ E E + R ++ E + +
Sbjct 431 LSMLIPSNMK----IQVVSQKFKGQEGNTNEPVYGTEIKVTRISSETMQKYEEALKTSHH 486
Query 107 AFRMPPPLLHIPKASELEILPG-LLGLNEPELISEQGGNAGTAVWWQGQGFSALPRVAVQ 165
A +P +I A++ + P L+ + P LI++ + + VW++ +P+ +
Sbjct 487 ALHLPEKNQYI--ATKFDQKPRELVKSDHPRLIND---DEWSRVWFKQDDEYKMPKQETK 541
Query 166 LSGSI----AKDKADLLSRTQGSVALAAIAEHLQEETADFQYCGITHSLAFKGTGFHMTF 221
L+ + + LLSR + L +++ L EE+ + G+ + L G M
Sbjct 542 LALTTPIVSQSPRMTLLSR----LWLRCLSDSLAEESYSAKVAGLNYELESSFFGVQMRV 597
Query 222 EGYTKTQLDKLMS-HVASVLRDPSVVESERFERVKQKQIKLLADPATNMAFEHALQAAAI 280
GY + Q L S H+ L + + + RF+ + + L + A + + + +
Sbjct 598 SGYAEKQ--ALFSKHLTKRLFNFKI-DQTRFDVLFDSLKRDLTNHAFSQPYVLSQHYTEL 654
Query 281 LTRNDAFSRMDLLNALEQTTYDDSIAKLSE--LKNVHVDAFVMGNIDRDQSLLLTESFLE 338
L + +S+ LL E +D + + + L+ H++ V GN +++ L++ ++
Sbjct 655 LVVDKEWSKQQLLAVCESVKLED-VQRFGKEMLQAFHLELLVYGNSTEKETIQLSKDLID 713
Query 339 -----QAGFTPIEHDDAVESLAMEQKQTIEATLANPIKGDKDHASLVQFQLGVPSIEERV 393
P+ ++ + ++ E + V +Q+GV + +
Sbjct 714 ILKSAAPSSRPLFRNEHILRREIQLNNGDEYIYRHLQTTHDVGCVQVTYQIGVQNTYDNA 773
Query 394 NLAILSQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGSKSHPDEVVKMIDA 453
+ ++ + +D+LRT+ LGYI + T LQ V+G KS D V++ I+A
Sbjct 774 VIGLIKNLITEPAFDTLRTKESLGYIVWTRTHFNCGTVALQILVQGPKS-VDHVLERIEA 832
Query 454 ELAKAKDYISNMPEAELSRWKEAAHAKLTKVEANFSEDFKKSADEIFSHANCFSKRDLEV 513
L + I MP+ E A+L + S FKK DEI F++ + +V
Sbjct 833 FLESVRKEIVEMPQEEFENRVSGLIAQLEEKPKTLSCRFKKFWDEIECRQYNFTRIEEDV 892
Query 514 KYL 516
+ L
Sbjct 893 ELL 895
> cel:C02G6.1 hypothetical protein; K01408 insulysin [EC:3.4.24.56]
Length=980
Score = 63.2 bits (152), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 91/446 (20%), Positives = 174/446 (39%), Gaps = 34/446 (7%)
Query 99 ERERERPNAFRMPPPLLHIPKASELEIL-----PGLLGLNE-PELISEQGGNAGTAVWWQ 152
E+ ++ NA + LH+P+ +E P NE P+LIS+ G + VW++
Sbjct 445 EKMKKYENALKTSHHALHLPEKNEYIATNFGQKPRESVKNEHPKLISDDGW---SRVWFK 501
Query 153 GQGFSALPRVAVQLSGSIAKDKADLLSRTQGSVALAAIAEHLQEETADFQYCGITHSLAF 212
+P+ + + + + S+ L + L EET + G+
Sbjct 502 QDDEYNMPKQETKFALTTPIVSQNPRISLISSLWLWCFCDILSEETYNAALAGLGCQFEL 561
Query 213 KGTGFH----------------MTFEGYTKTQLDKLMSHVASVLRDPSVVESERFERVKQ 256
G + GY + Q + H+ S + + + + RFE + +
Sbjct 562 SPFGVQKQSTDGREAERHASLTLHVYGYDEKQ-PLFVKHLTSCMINFKI-DRTRFEVLFE 619
Query 257 KQIKLLADPATNMAFEHALQAAAILTRNDAFSRMDLLNALEQTTYDDSIAKLSE-LKNVH 315
+ L + A + + +L + +S+ LL + T ++ E L+ H
Sbjct 620 SLKRTLTNHAFSQPYLLTQHYNQLLIVDKVWSKEQLLAVCDSVTLENVQGFAREMLQAFH 679
Query 316 VDAFVMGNIDRDQSLLLTESFLE-----QAGFTPIEHDDAVESLAMEQKQTIEATLANPI 370
++ FV GN +++ L++ ++ P+ ++ + E +
Sbjct 680 MELFVHGNSTEKEAIQLSKELMDILKSAAPNSRPLYRNEHNPRREFQLNNGDEYIYRHLQ 739
Query 371 KGDKDHASLVQFQLGVPSIEERVNLAILSQFLNRRIYDSLRTEAQLGYIAGAKESQAAST 430
K V +Q+GV + + + ++ Q + ++D+LRT LGYI
Sbjct 740 KTHDAGCVEVTYQIGVQNKYDNAVVGLIDQLIKEPVFDTLRTNEALGYIVWTGCRFNCGA 799
Query 431 ALLQCFVEGSKSHPDEVVKMIDAELAKAKDYISNMPEAELSRWKEAAHAKLTKVEANFSE 490
L FV+G KS D V++ I+ L + I MP+ E + A+L + S
Sbjct 800 VALNIFVQGPKS-VDYVLERIEVFLESVRKEIIEMPQDEFEKKVAGMIARLEEKPKTLSN 858
Query 491 DFKKSADEIFSHANCFSKRDLEVKYL 516
FK+ +I F++R+ EVK L
Sbjct 859 RFKRFWYQIECRQYDFARREKEVKVL 884
> sce:YLR389C STE23; Metalloprotease involved, with homolog Axl1p,
in N-terminal processing of pro-A-factor to the mature
form; member of the insulin-degrading enzyme family (EC:3.4.24.-);
K01408 insulysin [EC:3.4.24.56]
Length=1027
Score = 58.5 bits (140), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 83/452 (18%), Positives = 193/452 (42%), Gaps = 21/452 (4%)
Query 107 AFRMPPPLLHIPKASELEILPGLLGLNEPELISEQGGNAGTAVWWQGQGFSALPRVAVQL 166
A +P P + +++ + G+ L+EP L+ + + +W++ PR + L
Sbjct 530 ALTLPRPNEFVSTNFKVDKIDGIKPLDEPVLLL---SDDVSKLWYKKDDRFWQPRGYIYL 586
Query 167 SGSIAKDKADLLSRTQGSVALAAIAEHLQEETADFQYCGITHSLAFKGTGFHMTFEGYTK 226
S + A +++ ++ + L++ D + S G +T G+
Sbjct 587 SFKLPHTHASIINSMLSTLYTQLANDALKDVQYDAACADLRISFNKTNQGLAITASGFN- 645
Query 227 TQLDKLMSHVASVLRDPSVVE--SERFERVKQKQIKLLADPATNMAFEHALQAAAILTRN 284
+KL+ + L+ + E +RFE +K K I+ L + + + +
Sbjct 646 ---EKLIILLTRFLQGVNSFEPKKDRFEILKDKTIRHLKNLLYEVPYSQMSNYYNAIINE 702
Query 285 DAFSRMDLLNALEQTTYDDSIAKLSEL-KNVHVDAFVMGNIDRDQSL---LLTESFLEQA 340
++S + L E+ T++ I + + + V+ + + GNI +++L L +S +
Sbjct 703 RSWSTAEKLQVFEKLTFEQLINFIPTIYEGVYFETLIHGNIKHEEALEVDSLIKSLIPNN 762
Query 341 GFTPIEHDDAVESLAMEQKQTIEATLANPIKGDKDHASLVQF--QLGVPSIEERVNLAIL 398
++ + S + + +T A +K ++ S +Q QL V S + +
Sbjct 763 IHNLQVSNNRLRSYLLPKGKTFRYETA--LKDSQNVNSCIQHVTQLDVYSEDLSALSGLF 820
Query 399 SQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGSKSHPDEVVKMIDAELAKA 458
+Q ++ +D+LRT+ QLGY+ + TA ++ ++ + P + I+
Sbjct 821 AQLIHEPCFDTLRTKEQLGYVVFSSSLNNHGTANIRILIQSEHTTP-YLEWRINNFYETF 879
Query 459 KDYISNMPEAELSRWKEAAHAKLTKVEANFSEDFKKSADEIFSHANCFSKRDLEVKYLDN 518
+ +MPE + + KEA L + N +E+ + I+ F+ R + K + N
Sbjct 880 GQVLRDMPEEDFEKHKEALCNSLLQKFKNMAEESARYTAAIYLGDYNFTHRQKKAKLVAN 939
Query 519 DFSRKHLLRTFTK--LSDPSRRMVVKLIADLE 548
+++ ++ + +S+ + ++++ L + +E
Sbjct 940 -ITKQQMIDFYENYIMSENASKLILHLKSQVE 970
> mmu:230598 Nrd1, 2600011I06Rik, AI875733, MGC25477, NRD-C; nardilysin,
N-arginine dibasic convertase, NRD convertase 1 (EC:3.4.24.61);
K01411 nardilysin [EC:3.4.24.61]
Length=1161
Score = 52.4 bits (124), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 62/295 (21%), Positives = 126/295 (42%), Gaps = 13/295 (4%)
Query 252 ERVKQKQIKLLADPATNMAFEHALQAAAILTRNDAFSRMDLLNALEQTTYDDSIAK-LSE 310
E++K+ +L P T A ++ +S +D AL DS+ + +
Sbjct 807 EQLKKTYFNILIKPET-----LAKDVRLLILEYSRWSMIDKYQALMDGLSLDSLLNFVKD 861
Query 311 LKN-VHVDAFVMGNIDRDQSLLLTESFLEQAGFTPIEHDDAVESLAMEQKQTIEATLANP 369
K+ + V+ V GN+ +S+ + +++ F P+E + V+ +E
Sbjct 862 FKSQLFVEGLVQGNVTSTESMDFLKYVVDKLNFAPLEREMPVQFQVVELPSGHHLCKVRA 921
Query 370 I-KGDKDHASLVQFQLGVPSIEERVNLAILSQFLNRRIYDSLRTEAQLGY--IAGAKESQ 426
+ KGD + V +Q G S+ E + +L + +D LRT+ LGY + +
Sbjct 922 LNKGDANSEVTVYYQSGTRSLREYTLMELLVMHMEEPCFDFLRTKQTLGYHVYPTCRNTS 981
Query 427 AASTALLQCFVEGSKSHPDEVVKMIDAELAKAKDYISNMPEAELSRWKEAAHAKLTKVE- 485
+ + +K + + V K I+ L+ ++ I N+ E + + A KL + E
Sbjct 982 GILGFSVTVGTQATKYNSETVDKKIEEFLSSFEEKIENLTEDAFNT-QVTALIKLKECED 1040
Query 486 ANFSEDFKKSADEIFSHANCFSKRDLEVKYLDNDFSRKHLLRTFTKLSDPSRRMV 540
+ E+ ++ +E+ + F + E++ L + FS+ L+ F P +M+
Sbjct 1041 THLGEEVDRNWNEVVTQQYLFDRLAHEIEALKS-FSKSDLVSWFKAHRGPGSKML 1094
> hsa:4898 NRD1, hNRD1, hNRD2; nardilysin (N-arginine dibasic
convertase) (EC:3.4.24.61); K01411 nardilysin [EC:3.4.24.61]
Length=1151
Score = 52.4 bits (124), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 61/295 (20%), Positives = 128/295 (43%), Gaps = 13/295 (4%)
Query 252 ERVKQKQIKLLADPATNMAFEHALQAAAILTRNDAFSRMDLLNAL-EQTTYDDSIAKLSE 310
E++K+ +L P T A ++ +S +D AL + + + ++ + E
Sbjct 796 EQLKKTYFNILIKPET-----LAKDVRLLILEYARWSMIDKYQALMDGLSLESLLSFVKE 850
Query 311 LKN-VHVDAFVMGNIDRDQSLLLTESFLEQAGFTPIEHDDAVESLAMEQKQTIEATLANP 369
K+ + V+ V GN+ +S+ + +++ F P+E + V+ +E
Sbjct 851 FKSQLFVEGLVQGNVTSTESMDFLKYVVDKLNFKPLEQEMPVQFQVVELPSGHHLCKVKA 910
Query 370 I-KGDKDHASLVQFQLGVPSIEERVNLAILSQFLNRRIYDSLRTEAQLGY--IAGAKESQ 426
+ KGD + V +Q G S+ E + +L + +D LRT+ LGY + +
Sbjct 911 LNKGDANSEVTVYYQSGTRSLREYTLMELLVMHMEEPCFDFLRTKQTLGYHVYPTCRNTS 970
Query 427 AASTALLQCFVEGSKSHPDEVVKMIDAELAKAKDYISNMPEAELSRWKEAAHAKLTKVE- 485
+ + +K + + V K I+ L+ ++ I N+ E + + A KL + E
Sbjct 971 GILGFSVTVGTQATKYNSEVVDKKIEEFLSSFEEKIENLTEEAFNT-QVTALIKLKECED 1029
Query 486 ANFSEDFKKSADEIFSHANCFSKRDLEVKYLDNDFSRKHLLRTFTKLSDPSRRMV 540
+ E+ ++ +E+ + F + E++ L + FS+ L+ F P +M+
Sbjct 1030 THLGEEVDRNWNEVVTQQYLFDRLAHEIEALKS-FSKSDLVNWFKAHRGPGSKML 1083
> tgo:TGME49_044490 insulin-degrading enzyme, putative (EC:3.4.24.56)
Length=592
Score = 52.0 bits (123), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 57/268 (21%), Positives = 106/268 (39%), Gaps = 44/268 (16%)
Query 286 AFSRMDLLNALEQTTYDDSIAKLSELKNVHVDAFVMGNIDRDQSLLLTESFLEQAGFTPI 345
S DLL LEQTT D + + V+ V+GNI + ++ E L+
Sbjct 193 CLSYEDLLRVLEQTTLDVQEVPKTLFERACVEGLVVGNISSAEVCVMVEMALKNLNIETT 252
Query 346 EHDDAV-----------------------------------ESLAMEQKQTIEATLA--- 367
++V +L E+ + E L
Sbjct 253 LDSNSVPEKAVVDLASLDLARLRSSGSGVAGLDEEREAMQCRTLVCEELKASEVKLKTRS 312
Query 368 -NPIKGDKDHASLVQFQLGVPSIEERVNLAILSQFLNRRIYDSLRTEAQLGYIAGAKESQ 426
N D++ + ++FQLG ER L++ S +++ +D LRT+ QL Y+ A S
Sbjct 313 ENSNTQDRNSVAFLRFQLGNLEDRERSMLSLFSHCISQAFFDDLRTQQQLDYVLHAHRSF 372
Query 427 AASTALLQCFVEGSKSHPD----EVVKMIDAELAKAKDYISNMPEAELSRWKEAAHAKLT 482
+ + FV GS + D + ++++ L+ + + +P+A + + A ++L
Sbjct 373 QLRSQGMHFFVAGS-TFSDLMTLRIGRLVEKYLSSEQGLHAGLPDALCEKHRSALVSELR 431
Query 483 KVEANFSEDFKKSADEIFSHANCFSKRD 510
N E+ ++ EI + F++ D
Sbjct 432 VRPQNAFEEAQRYTREISTWYFMFNRHD 459
> tgo:TGME49_069870 hypothetical protein
Length=413
Score = 50.4 bits (119), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 39/149 (26%), Positives = 78/149 (52%), Gaps = 19/149 (12%)
Query 358 QKQTIEATLANPIKGDKDHASLVQFQLG-VPSIEERVNLAILSQFLNRRIYDSLRTEAQL 416
Q ++I L NP DK + + + ++G +P+I +R L ++S+++++R ++ LRTE QL
Sbjct 190 QLRSIRKNL-NP--NDKKNQAYLLIEVGALPNIHDRAVLYMVSRWMSQRFFNKLRTEQQL 246
Query 417 GYIAGAKESQAASTALLQCFVEGSKSHP----DEVVKMIDAELAK--AKDYISNMPEAEL 470
GY+ S+ + F+ S P D +V+ I+AE +K ++ + + +A +
Sbjct 247 GYLTAMHSSRLEDRFYYRFFIT-STYDPAEVADRIVEFINAERSKIPTQEEFATLKQAAI 305
Query 471 SRWKEAAHAKLTKVEANFSEDFKKSADEI 499
WK+ N E+F+K+ ++
Sbjct 306 DVWKQKPK--------NIFEEFRKNRRQV 326
> cel:C28F5.4 hypothetical protein; K01408 insulysin [EC:3.4.24.56]
Length=856
Score = 48.1 bits (113), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 86/410 (20%), Positives = 157/410 (38%), Gaps = 35/410 (8%)
Query 135 PELISEQGGNAGTAVWWQGQGFSALPRVAVQLSGSIAKDKADLLSRTQGSVALAAIAEHL 194
P LISE + VW++ P+ + + + L+++ +V + +
Sbjct 450 PRLISE---DEWIQVWFKQDNEYNSPKQGIMFALTTP-----LVAKKSKNVVAFKSLDTI 501
Query 195 QEETADFQYCGITHSLAFKGTGFHMTFEGYTKTQLDKLMSHVASVLRDPSV---VESERF 251
EET + + G+ +G + GY + Q H+ + + + V F
Sbjct 502 IEETYNARLAGLECQFESSSSGVQIRVFGYDEKQ-SLFAKHLVNRMANFQVNRLCFDISF 560
Query 252 ERVKQKQIKLLADPATNMAFE--HALQAAAI--LTRNDAFSRMDLLNALEQTTYDDSIA- 306
E +K+ TN AF H L A I L ++ +S+ LL + T +D
Sbjct 561 ESLKRT--------LTNHAFSQPHDLSAHFIDLLVVDNIWSKEQLLAVCDSVTLEDVHGF 612
Query 307 KLSELKNVHVDAFVMGNIDRDQSLLLTESFLE-----QAGFTPIEHDDAVESLAMEQKQT 361
+ L+ H++ FV GN +L L++ + P++ D+ ++
Sbjct 613 AIKMLQAFHMELFVHGNSTEKDTLQLSKELSDILKSVAPNSRPLKRDEHNPHRELQLING 672
Query 362 IEATLANPIKGDKDHASLVQFQLGVPSIEERVNLAILSQFLNRRIYDSLRTEAQLGYIAG 421
E + K V FQ+GV S +L++ + Y LRT LGY
Sbjct 673 HEHVYRHFQKTHDVGCVEVAFQIGVQSTYNNSVNKLLNELIKNPAYTILRTNEALGYNVS 732
Query 422 AKESQAASTALLQCFVEGSKSHPDEVVKMIDAELAKAKDYISNMPEAELSRWKEAAHAKL 481
+ L V+G +S D V++ I+ L A++ I MP+ + A
Sbjct 733 TESRLNDGNVYLHVIVQGPES-ADHVLERIEVFLESAREEIVAMPQEDFDY---QVWAMF 788
Query 482 TKVEANFSEDFKKSADEIFSHANCFSKRDLEVKYLDNDFSRKHLLRTFTK 531
+ S+ F EI S F R+ EV+ + +++ ++ F +
Sbjct 789 KENPPTLSQCFSMFWSEIHSRQYNFG-RNKEVRGISKRITKEEVINFFDR 837
> ath:AT2G41790 peptidase M16 family protein / insulinase family
protein; K01408 insulysin [EC:3.4.24.56]
Length=970
Score = 44.7 bits (104), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 67/347 (19%), Positives = 139/347 (40%), Gaps = 33/347 (9%)
Query 190 IAEHLQEETADFQYCGITHSLAFKGTGFHMTFEGYT---KTQLDKLMSHVASVLRDPSVV 246
+ ++L E Q G+ + ++ GF +T GY + L+ ++ +A+ P
Sbjct 560 LMDYLNEYAYYAQVAGLYYGVSLSDNGFELTLLGYNHKLRILLETVVGKIANFEVKP--- 616
Query 247 ESERFERVKQKQIKLLADPATNMAFEHALQAAAILTRNDAFSRMDLLNALEQTTYDDSIA 306
+RF +K+ K + + A+ +++ ++ + + L+ L +D +A
Sbjct 617 --DRFAVIKETVTKEYQNYKFRQPYHQAMYYCSLILQDQTWPWTEELDVLSHLEAED-VA 673
Query 307 KLSE--LKNVHVDAFVMGNIDRDQSLLLTESFLEQAGFT----------PIEH-DDAVES 353
K L ++ ++ GN++ +++ + + +E F P +H + V
Sbjct 674 KFVPMLLSRTFIECYIAGNVENNEAESMVKH-IEDVLFNDPKPICRPLFPSQHLTNRVVK 732
Query 354 LAMEQKQTIEATLANPIKGDKDHASLVQFQLGVPSIEERVNLAILSQFLNRRIYDSLRTE 413
L K +NP D++ A + Q+ + L + + + LRT
Sbjct 733 LGEGMKYFYHQDGSNP--SDENSALVHYIQVHRDDFSMNIKLQLFGLVAKQATFHQLRTV 790
Query 414 AQLGYIAGAKESQAASTALLQCFVEGSKSHPDEVVKMIDAELAKAKDYISNMPEAELSRW 473
QLGYI + + +Q ++ S P + +++ L K++ S + E +
Sbjct 791 EQLGYITALAQRNDSGIYGVQFIIQSSVKGPGHIDSRVESLL---KNFESKLYEMSNEDF 847
Query 474 KEAAHA----KLTKVEANFSEDFKKSADEIFSHANCFSKRDLEVKYL 516
K A KL K N E+ + EI S F++++ EV L
Sbjct 848 KSNVTALIDMKLEK-HKNLKEESRFYWREIQSGTLKFNRKEAEVSAL 893
> dre:557565 nrd1, si:dkey-171o17.4; nardilysin (N-arginine dibasic
convertase) (EC:3.4.24.61)
Length=1061
Score = 42.7 bits (99), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 33/136 (24%), Positives = 56/136 (41%), Gaps = 6/136 (4%)
Query 289 RMDLLNALEQTTYDDSIAKLSELKN-----VHVDAFVMGNIDRDQSLLLTESFLEQAGFT 343
R ++ E D S+A L N + V+ V GN +S + F+E+ +
Sbjct 733 RWSVMQKYEAIMADPSVADLMTFANRFKAELFVEGLVQGNFTSAESKEFLQCFIEKLKYA 792
Query 344 PIEHDDAVESLAMEQKQTIEATLANPI-KGDKDHASLVQFQLGVPSIEERVNLAILSQFL 402
P + V +E QT + K D + V +Q G+ ++ E + +L +
Sbjct 793 PHPIEPPVLFRVVELPQTHHLCKVQSLNKADANSEVTVYYQTGLKNLREHTLMELLVMHM 852
Query 403 NRRIYDSLRTEAQLGY 418
+D LRT+ LGY
Sbjct 853 EEPCFDFLRTKETLGY 868
> ath:AT1G06900 catalytic/ metal ion binding / metalloendopeptidase/
zinc ion binding; K01411 nardilysin [EC:3.4.24.61]
Length=1024
Score = 42.4 bits (98), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 82/422 (19%), Positives = 172/422 (40%), Gaps = 50/422 (11%)
Query 150 WWQGQGFSALPRVA----VQLSGSIAKDKADLLSRTQGSVALAAIAEHLQEETADFQYCG 205
W++ +PR + L G+ A K LL+ + + + + L E
Sbjct 588 WYKLDETFKVPRANTYFRINLKGAYASVKNCLLTE----LYINLLKDELNEIIYQASIAK 643
Query 206 ITHSLAFKGTGFHMTFEGYTKTQLDKLMSHVASVLRD--PSVVESERFERVKQKQIKLLA 263
+ SL+ G + G+ + ++ L+S + ++ + P++ ERF+ +K+ +
Sbjct 644 LETSLSMYGDKLELKVYGFNE-KIPALLSKILAIAKSFMPNL---ERFKVIKENMERGFR 699
Query 264 DPATNMA-FEHALQAAAILTRNDAFSRMDLLNALEQTTYDDSIAKLSELKN-VHVDAFVM 321
+ TNM H+ L + + L+ L + DD + + EL++ + ++A
Sbjct 700 N--TNMKPLNHSTYLRLQLLCKRIYDSDEKLSVLNDLSLDDLNSFIPELRSQIFIEALCH 757
Query 322 GNIDRDQSLLLTESFLEQAGFTPI----EHDDAVESLAMEQKQTIEATLANPIKGDKDHA 377
GN+ D+++ ++ F + P+ H + + M K + + N K + +
Sbjct 758 GNLSEDEAVNISNIFKDSLTVEPLPSKCRHGEQITCFPMGAKLVRDVNVKN--KSETNSV 815
Query 378 SLVQFQLG---VPSIEERVNLAILSQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQ 434
+ +Q+ S + L + + + +++ LRT+ QLGY+
Sbjct 816 VELYYQIEPEEAQSTRTKAVLDLFHEIIEEPLFNQLRTKEQLGYVVECGPRLTYRVHGFC 875
Query 435 CFVEGSKSHP-------DEVVKMIDAELAK-----AKDYISNMPEAELSRWKEAAHAKLT 482
V+ SK P D +K I+ L + +DY S M ++R E + L+
Sbjct 876 FCVQSSKYGPVHLLGRVDNFIKDIEGLLEQLDDESYEDYRSGM----IARLLEKDPSLLS 931
Query 483 KVEANFSEDFKKSADEIFSHANCFSKRDLEVKYLDNDFSRKHLLRTFTKLSDP-SRRMVV 541
+ +S+ K FSH R ++ K + + + +T+ + S P RR+ V
Sbjct 932 ETNDLWSQIVDKRYMFDFSHKEAEELRSIQKKDVISWY------KTYFRESSPKCRRLAV 985
Query 542 KL 543
++
Sbjct 986 RV 987
> ath:AT3G57470 peptidase M16 family protein / insulinase family
protein; K01408 insulysin [EC:3.4.24.56]
Length=891
Score = 41.2 bits (95), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 56/302 (18%), Positives = 116/302 (38%), Gaps = 25/302 (8%)
Query 185 VALAAIAEHLQEETADFQYCGITHSLAFKGTGFHMTFEGYT---KTQLDKLMSHVASVLR 241
+ + + ++L E Q G+ + L+ GF ++ G+ + L+ ++ +A
Sbjct 482 IFVWLLVDYLNEYAYYAQAAGLDYGLSLSDNGFELSLAGFNHKLRILLEAVIQKIAKFEV 541
Query 242 DPSVVESERFERVKQKQIKLLADPATNMAFEHALQAAAILTRNDAFSRMDLLNALEQTTY 301
P +RF +K+ K + E A +++ ++ + + L+AL
Sbjct 542 KP-----DRFSVIKETVTKAYQNNKFQQPHEQATNYCSLVLQDQIWPWTEELDALSHLEA 596
Query 302 DDSIAKLSE--LKNVHVDAFVMGNIDRDQSLLLTESFLEQAGFT---PIEH--------D 348
+D +A L V+ ++ GN+++D++ + + +E FT PI
Sbjct 597 ED-LANFVPMLLSRTFVECYIAGNVEKDEAESMVKH-IEDVLFTDSKPICRPLFPSQFLT 654
Query 349 DAVESLAMEQKQTIEATLANPIKGDKDHASLVQFQLGVPSIEERVNLAILSQFLNRRIYD 408
+ V L K +N D++ A + Q+ L + + +
Sbjct 655 NRVTELGTGMKHFYYQEGSN--SSDENSALVHYIQVHKDEFSMNSKLQLFELIAKQDTFH 712
Query 409 SLRTEAQLGYIAGAKESQAASTALLQCFVEGSKSHPDEVVKMIDAELAKAKDYISNMPEA 468
LRT QLGYI S + +Q ++ S P + +++ L + NM +
Sbjct 713 QLRTIEQLGYITSLSLSNDSGVYGVQFIIQSSVKGPGHIDSRVESLLKDLESKFYNMSDE 772
Query 469 EL 470
E
Sbjct 773 EF 774
> dre:393572 traf3ip1, Elipsa, MGC63522, zgc:63522; TNF receptor-associated
factor 3 interacting protein 1
Length=629
Score = 41.2 bits (95), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 21/39 (53%), Positives = 34/39 (87%), Gaps = 1/39 (2%)
Query 66 TRRRNKTKNSRERERERERERERERERERERERERERER 104
+R R K K +RE+ERERE++R RE+ERER+++R++++ER
Sbjct 219 SRDREKDK-TREKEREREKDRNREKERERDKDRDKKKER 256
Score = 40.0 bits (92), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 21/39 (53%), Positives = 33/39 (84%), Gaps = 1/39 (2%)
Query 67 RRRNKTKN-SRERERERERERERERERERERERERERER 104
R R+K K+ RE+++ RE+ERERE++R RE+ERER+++R
Sbjct 212 RDRDKDKSRDREKDKTREKEREREKDRNREKERERDKDR 250
Score = 38.9 bits (89), Expect = 0.056, Method: Compositional matrix adjust.
Identities = 19/37 (51%), Positives = 33/37 (89%), Gaps = 1/37 (2%)
Query 67 RRRNKTKNSRERERERERERERERERERERERERERE 103
R ++KT+ +ERERE++R RE+ERER+++R++++ERE
Sbjct 222 REKDKTRE-KEREREKDRNREKERERDKDRDKKKERE 257
Score = 37.0 bits (84), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 19/41 (46%), Positives = 32/41 (78%), Gaps = 3/41 (7%)
Query 67 RRRNKTKN---SRERERERERERERERERERERERERERER 104
R R +TK+ +++ R+RE+++ RE+ERERE++R RE+ER
Sbjct 204 RERERTKDRDRDKDKSRDREKDKTREKEREREKDRNREKER 244
> eco:b2821 ptrA, ECK2817, JW2789, ptr; protease III (EC:3.4.24.55);
K01407 protease III [EC:3.4.24.55]
Length=962
Score = 38.5 bits (88), Expect = 0.072, Method: Compositional matrix adjust.
Identities = 69/313 (22%), Positives = 125/313 (39%), Gaps = 19/313 (6%)
Query 205 GITHSLAFKGTGFHMTFEGYTKTQLDKLMSHVASVLRDPSVVESERFERVKQKQIKLLAD 264
GI+ S G + GYT+ +L +L + + E ++ E+ K +++
Sbjct 591 GISFSTN-ANNGLMVNANGYTQ-RLPQLFQALLEGYFSYTATE-DQLEQAKSWYNQMMDS 647
Query 265 PATNMAFEHALQAAAILTRNDAFSRMDLLNALEQTTYDDSIAKLSELKNVHVDAF-VMGN 323
AFE A+ A +L++ FSR + L T + +A LK+ F V+GN
Sbjct 648 AEKGKAFEQAIMPAQMLSQVPYFSRDERRKILPSITLKEVLAYRDALKSGARPEFMVIGN 707
Query 324 IDRDQSLLLTESFLEQAGFTPIE---HDDAVESLAMEQKQTIEATLANPIKGDKDHASL- 379
+ Q+ L +Q G E + D V +++KQ++ A G+ ++L
Sbjct 708 MTEAQATTLARDVQKQLGADGSEWCRNKDVV----VDKKQSVIFEKA----GNSTDSALA 759
Query 380 -VQFQLGVPSIEERVNLAILSQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVE 438
V G ++L Q + Y+ LRTE QLGY A + ++
Sbjct 760 AVFVPTGYDEYTSSAYSSLLGQIVQPWFYNQLRTEEQLGYAVFAFPMSVGRQWGMGFLLQ 819
Query 439 GSKSHPDEVVKMIDAELAKAKDYISNMPEAELSRWKEAAHAKLTKVEANFSEDFKKSADE 498
+ P + + A A+ + M E ++ ++A ++ + E+ K + +
Sbjct 820 SNDKQPSFLWERYKAFFPTAEAKLRAMKPDEFAQIQQAVITQMLQAPQTLGEEASKLSKD 879
Query 499 IFSHANC-FSKRD 510
F N F RD
Sbjct 880 -FDRGNMRFDSRD 891
> cpv:cgd2_920 peptidase'insulinase-like peptidase'
Length=1028
Score = 37.7 bits (86), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 45/243 (18%), Positives = 111/243 (45%), Gaps = 11/243 (4%)
Query 287 FSRMDLLNALEQTTYDDSIA-KLSELKNVHVDAFVMGNIDRDQSLLLT-ESFLEQAGF-T 343
F+R + LN LE T++ + + L N ++ +MGN + ++ + + F
Sbjct 711 FNRQEKLNVLESFTFELFCSIRQHFLSNCRLEGLIMGNFSEPNAKCISIQHWKNLINFQN 770
Query 344 PIEHDDAVESLAMEQ------KQTIEATLANPIKGDKDHASLVQFQLGVPSIEERVNLAI 397
++++ + +EQ K+ I P DK+ ++ F LG ++ ++V +
Sbjct 771 SVKNEVKSCGIKVEQFSIVNLKKDIYTLNYIPNSSDKNGCWMLSFFLGEYNLRKQVLCDL 830
Query 398 LSQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGSK-SHPDEVVKMIDAELA 456
+ F++ + LRT QL Y+ A + ++ ++ +++ S+ ++ + ++++ +
Sbjct 831 ILPFVSSEAFADLRTNQQLAYVVRATQIFSSPAIIIGYYLQSSEYTNALTLERLLEFHIN 890
Query 457 KAK-DYISNMPEAELSRWKEAAHAKLTKVEANFSEDFKKSADEIFSHANCFSKRDLEVKY 515
K K + S + + + K++ L+ + +++K EI + F R ++
Sbjct 891 KTKVELKSKLNKEMFIKLKDSTIQTLSSNPKSIFDEYKTYLHEINERSYLFDIRQRKIDI 950
Query 516 LDN 518
L+N
Sbjct 951 LNN 953
> mmu:209361 Taf3, 140kDa, 4933439M23Rik, AW539625, TAF140, TAFII-140,
TAFII140, mTAFII140; TAF3 RNA polymerase II, TATA box
binding protein (TBP)-associated factor; K14650 transcription
initiation factor TFIID subunit 3
Length=932
Score = 36.6 bits (83), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 32/47 (68%), Gaps = 4/47 (8%)
Query 76 RERERERERERERERER----ERERERERERERPNAFRMPPPLLHIP 118
RERE+ ++++++RER + +RERER +E+ R + + PP L +P
Sbjct 613 REREKHKDKKKDRERSKREKDKRERERLKEKNREDKIKAPPTQLVLP 659
Score = 32.7 bits (73), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 39/75 (52%), Gaps = 4/75 (5%)
Query 67 RRRNKTKNSRERERERERERERERERERERERERE---RERPNAFRMPPPLLHIPKASEL 123
RR + ++++RER + + +RERER +E+ RE + P +PP + +P S
Sbjct 612 RREREKHKDKKKDRERSKREKDKRERERLKEKNREDKIKAPPTQLVLPPKEMALPLFSPS 671
Query 124 EI-LPGLLGLNEPEL 137
+ +P +L P L
Sbjct 672 AVRVPAMLPAFSPML 686
> pfa:PFE0230w conserved Plasmodium protein, unknown function
Length=2349
Score = 35.8 bits (81), Expect = 0.56, Method: Composition-based stats.
Identities = 17/33 (51%), Positives = 25/33 (75%), Gaps = 1/33 (3%)
Query 76 RERERERERERERERERERERER-ERERERPNA 107
+E+ER +ERE RE+ER RE+ER ++ER + A
Sbjct 1461 KEKERYKEREILREKERLREKERLKQERLKKEA 1493
> tgo:TGME49_016980 hypothetical protein
Length=530
Score = 35.4 bits (80), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 16/31 (51%), Positives = 26/31 (83%), Gaps = 0/31 (0%)
Query 74 NSRERERERERERERERERERERERERERER 104
N +ER+RE+E E+ER R R+RE +++R+RE+
Sbjct 411 NKKERKREQEGEKERTRRRDRENKKKRQREQ 441
> ath:AT5G67320 HOS15; HOS15 (high expression of osmotically responsive
genes 15)
Length=613
Score = 34.3 bits (77), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 14/42 (33%), Positives = 27/42 (64%), Gaps = 0/42 (0%)
Query 63 LLSTRRRNKTKNSRERERERERERERERERERERERERERER 104
+L ++R + +ER+R +E ++ ERE E +R R +E++R
Sbjct 97 MLREKKRKERDMEKERDRSKENDKGVEREHEGDRNRAKEKDR 138
Score = 31.6 bits (70), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 13/32 (40%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 76 RERERERERERERERERERERERERERERPNA 107
+ +ER+ E+ER+R +E ++ ERE E +R A
Sbjct 102 KRKERDMEKERDRSKENDKGVEREHEGDRNRA 133
> pfa:MAL7P1.129 conserved Plasmodium protein, unknown function
Length=1003
Score = 32.3 bits (72), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 17/29 (58%), Positives = 24/29 (82%), Gaps = 0/29 (0%)
Query 75 SRERERERERERERERERERERERERERE 103
+RERE+E +RER+ +R RE+ER +RERE
Sbjct 664 NREREKEMDRERDMDRNREKERNMDRERE 692
> mmu:226562 Prrc2c, 1810043M20Rik, 9630039I18Rik, A630006J20,
Bat2d, Bat2d1, Bat2l2, E130112L15Rik, Prrc3, mKIAA1096; proline-rich
coiled-coil 2C
Length=2846
Score = 32.3 bits (72), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 15/29 (51%), Positives = 24/29 (82%), Gaps = 0/29 (0%)
Query 76 RERERERERERERERERERERERERERER 104
RE ER R++E+E E++RE+E+E +R RE+
Sbjct 534 REMERARQQEKELEQQREKEQELQRLREQ 562
Lambda K H
0.316 0.132 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 31498167456
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40