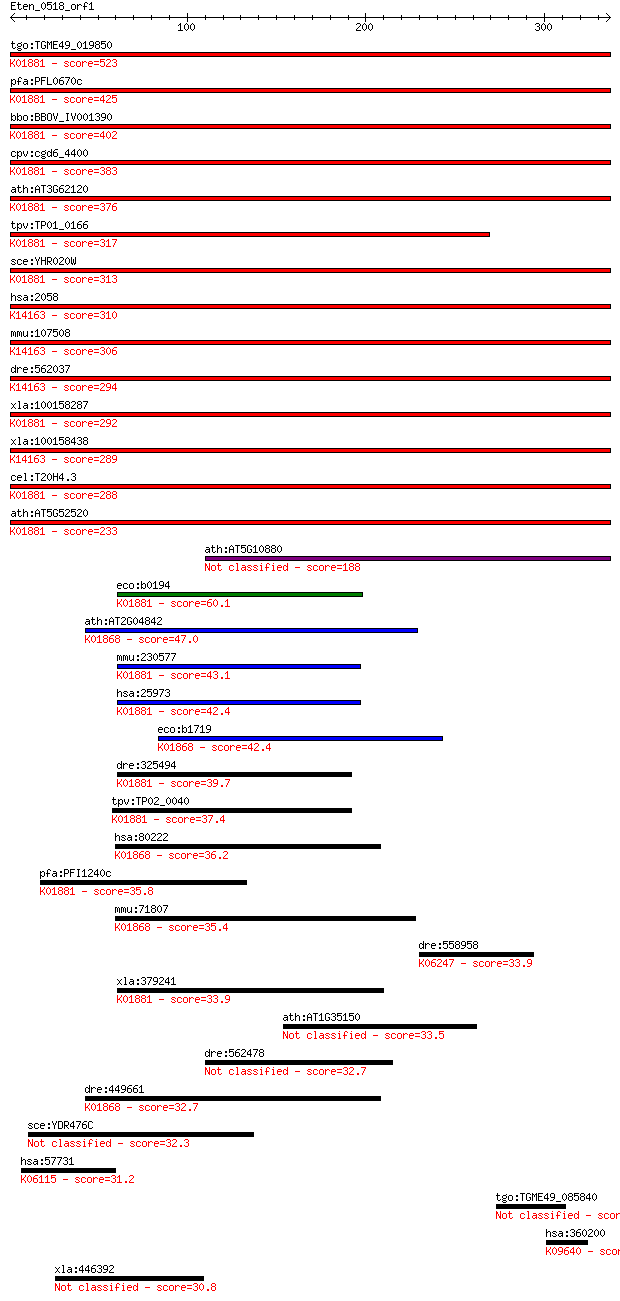
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0518_orf1
Length=336
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_019850 prolyl-tRNA synthetase, putative (EC:6.1.1.1... 523 4e-148
pfa:PFL0670c bifunctional aminoacyl-tRNA synthetase, putative ... 425 1e-118
bbo:BBOV_IV001390 21.m02889; prolyl-tRNA synthetase (EC:6.1.1.... 402 9e-112
cpv:cgd6_4400 proline-tRNA synthetase; class II aaRS (ybak RNA... 383 4e-106
ath:AT3G62120 tRNA synthetase class II (G, H, P and S) family ... 376 8e-104
tpv:TP01_0166 prolyl-tRNA synthetase; K01881 prolyl-tRNA synth... 317 4e-86
sce:YHR020W Protein of unknown function that may interact with... 313 7e-85
hsa:2058 EPRS, DKFZp313B047, EARS, GLUPRORS, PARS, QARS, QPRS;... 310 5e-84
mmu:107508 Eprs, 2410081F06Rik, 3010002K18Rik, C79379, Qprs; g... 306 6e-83
dre:562037 eprs, wu:fb38d08, wu:ft34d10; glutamyl-prolyl-tRNA ... 294 4e-79
xla:100158287 hypothetical protein LOC100158287; K01881 prolyl... 292 9e-79
xla:100158438 eprs; glutamyl-prolyl-tRNA synthetase; K14163 bi... 289 1e-77
cel:T20H4.3 prs-1; Prolyl tRNA Synthetase family member (prs-1... 288 3e-77
ath:AT5G52520 OVA6; OVA6 (OVULE ABORTION 6); ATP binding / ami... 233 6e-61
ath:AT5G10880 tRNA synthetase-related / tRNA ligase-related 188 3e-47
eco:b0194 proS, drpA, ECK0194, JW0190; prolyl-tRNA synthetase ... 60.1 1e-08
ath:AT2G04842 EMB2761 (EMBRYO DEFECTIVE 2761); ATP binding / a... 47.0 1e-04
mmu:230577 Pars2, BC027073, MGC47008; prolyl-tRNA synthetase (... 43.1 0.002
hsa:25973 PARS2, DKFZp727A071, MGC14416, MGC19467, MT-PRORS; p... 42.4 0.002
eco:b1719 thrS, ECK1717, JW1709; threonyl-tRNA synthetase (EC:... 42.4 0.002
dre:325494 pars2, fc83e06, wu:fc83e06, zgc:77163; prolyl-tRNA ... 39.7 0.018
tpv:TP02_0040 prolyl-tRNA synthetase; K01881 prolyl-tRNA synth... 37.4 0.072
hsa:80222 TARS2, FLJ12528, TARSL1; threonyl-tRNA synthetase 2,... 36.2 0.20
pfa:PFI1240c prolyl-t-RNA synthase, putative (EC:6.1.1.15); K0... 35.8 0.22
mmu:71807 Tars2, 2610024N01Rik, AI429208, Tarsl1; threonyl-tRN... 35.4 0.35
dre:558958 lamc3, si:dkey-245l10.1; laminin, gamma 3; K06247 l... 33.9 0.94
xla:379241 pars2, MGC53152; prolyl-tRNA synthetase 2, mitochon... 33.9 0.96
ath:AT1G35150 hypothetical protein 33.5 1.2
dre:562478 zufsp, MGC158611, im:7145879, zgc:158611; zinc fing... 32.7 1.8
dre:449661 tars, ThrRS, wu:fb07c05, wu:fb39c12, zgc:92586; thr... 32.7 1.9
sce:YDR476C Putative protein of unknown function; green fluore... 32.3 2.8
hsa:57731 SPTBN4, KIAA1642, QV, SPNB4, SPTBN3; spectrin, beta,... 31.2 5.5
tgo:TGME49_085840 hypothetical protein 30.8 7.3
hsa:360200 TMPRSS9, FLJ16193; transmembrane protease, serine 9... 30.8 7.6
xla:446392 b9d1, MGC83744; B9 protein domain 1 30.8
> tgo:TGME49_019850 prolyl-tRNA synthetase, putative (EC:6.1.1.15);
K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=711
Score = 523 bits (1347), Expect = 4e-148, Method: Compositional matrix adjust.
Identities = 242/339 (71%), Positives = 287/339 (84%), Gaps = 3/339 (0%)
Query 1 TAHATEEEAYTLVLEILELYRQWYEDYLAVPVIKGEKSENEKFAGGKKTTTIEGIIPDTG 60
TAHATEEEA+ LVL+ILELYR+WYE+ LAVPVIKGEKSE EKFAGGKKTTT+E IP+ G
Sbjct 373 TAHATEEEAWELVLDILELYRRWYEECLAVPVIKGEKSEGEKFAGGKKTTTVEAFIPENG 432
Query 61 RGIQAATSHLLGQNFSRMFSIEFEDEKGAKQLVHQTSWGCTTRSLGVMIMTHGDDKGLVL 120
RGIQAATSHLLG NF++MF IEFEDE+G K+LVHQTSWGCTTRSLGVMIMTHGDDKGLV+
Sbjct 433 RGIQAATSHLLGTNFAKMFEIEFEDEEGHKRLVHQTSWGCTTRSLGVMIMTHGDDKGLVI 492
Query 121 PPRVVAVQAVIIPIIFKETGGMEIVAKCRELERLLNAAGVRVKVDDRTNYTPGWKFNDWE 180
PPRV +VQ VIIPI+FK+ EI+ KCREL+ +L A +RV++DDR+NYTPGWK+N WE
Sbjct 493 PPRVASVQVVIIPILFKDENTGEILGKCRELKTMLEKADIRVRIDDRSNYTPGWKYNHWE 552
Query 181 LKGVPLRLEIGPRDVESCQTRVVRRDTSEARNVPWAEAASTIPAMLETMQKDLFNKAKAK 240
+KGVPLRLE+GP+D+ RVVRRDT EA + WA+ A + ++E +Q+ LF KAKA+
Sbjct 553 VKGVPLRLELGPKDLAKGTARVVRRDTGEAYQISWADLAPKLLELMEGIQRSLFEKAKAR 612
Query 241 FEASIEQITTFDEVMPALNRRHVVLAPWCEDPETETQIKRETQRLSEIQARENADG---M 297
IE+I+TFDEVMPALNR+H+VLAPWCEDPE+E QIK+ETQ+LSEIQA E D M
Sbjct 613 LHEGIEKISTFDEVMPALNRKHLVLAPWCEDPESEEQIKKETQKLSEIQAIEAGDSEQVM 672
Query 298 TGAMKPLCIPFDQPPMPEGTRCFFTGRPAKRWCLFGRSY 336
TGAMK LCIPFDQPPMPEGT+CF+TG+PAKRW L+GRSY
Sbjct 673 TGAMKTLCIPFDQPPMPEGTKCFYTGKPAKRWTLWGRSY 711
> pfa:PFL0670c bifunctional aminoacyl-tRNA synthetase, putative
(EC:6.1.1.15); K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=746
Score = 425 bits (1092), Expect = 1e-118, Method: Compositional matrix adjust.
Identities = 193/336 (57%), Positives = 246/336 (73%), Gaps = 1/336 (0%)
Query 1 TAHATEEEAYTLVLEILELYRQWYEDYLAVPVIKGEKSENEKFAGGKKTTTIEGIIPDTG 60
TAH EEEA LV +IL+LYR+WYE+YLAVP+IKG KSE EKF G T+T E I + G
Sbjct 412 TAHKNEEEAVKLVFDILDLYRRWYEEYLAVPIIKGIKSEGEKFGGANFTSTAEAFISENG 471
Query 61 RGIQAATSHLLGQNFSRMFSIEFEDEKGAKQLVHQTSWGCTTRSLGVMIMTHGDDKGLVL 120
R IQAATSH LG NF++MF IEFEDE KQ VHQTSWGCTTRS+G+MIMTHGDDKGLVL
Sbjct 472 RAIQAATSHYLGTNFAKMFKIEFEDENEVKQYVHQTSWGCTTRSIGIMIMTHGDDKGLVL 531
Query 121 PPRVVAVQAVIIPIIFKETGGMEIVAKCRELERLLNAAGVRVKVDDRTNYTPGWKFNDWE 180
PP V + VI+PI +K T I + C+++E++L A + DDR +Y+PG+KFN WE
Sbjct 532 PPNVSKYKVVIVPIFYKTTDENAIHSYCKDIEKILKNAQINCVYDDRASYSPGYKFNHWE 591
Query 181 LKGVPLRLEIGPRDVESCQTRVVRRDTSEARNVPWAEAASTIPAMLETMQKDLFNKAKAK 240
L+G+P+R+E+GP+D+++ +VRRD +E NV ML + K+LF KAK K
Sbjct 592 LRGIPIRIEVGPKDLQNNSCVIVRRDNNEKCNVKKESVLLETQQMLVDIHKNLFLKAKKK 651
Query 241 FEASIEQITTFDEVMPALNRRHVVLAPWCEDPETETQIKRETQRLSEIQARENADGMTGA 300
+ SI Q+T+F EVM ALN++ +VLAPWCED TE +IK+ETQRLS + + ++GA
Sbjct 652 LDDSIVQVTSFSEVMNALNKKKMVLAPWCEDIATEEEIKKETQRLS-LNQTNSETTLSGA 710
Query 301 MKPLCIPFDQPPMPEGTRCFFTGRPAKRWCLFGRSY 336
MKPLCIP DQPPMP +CF++G+PAKRWCLFGRSY
Sbjct 711 MKPLCIPLDQPPMPPNMKCFWSGKPAKRWCLFGRSY 746
> bbo:BBOV_IV001390 21.m02889; prolyl-tRNA synthetase (EC:6.1.1.15);
K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=705
Score = 402 bits (1033), Expect = 9e-112, Method: Compositional matrix adjust.
Identities = 192/336 (57%), Positives = 244/336 (72%), Gaps = 4/336 (1%)
Query 1 TAHATEEEAYTLVLEILELYRQWYEDYLAVPVIKGEKSENEKFAGGKKTTTIEGIIPDTG 60
TAH +EE+A V+ +L LY+++YE++LAVPVI GEKS E+FAGGK T TIE IP +G
Sbjct 374 TAHKSEEDAMNTVMTMLRLYQRFYEEFLAVPVIPGEKSVEERFAGGKSTMTIEAYIPGSG 433
Query 61 RGIQAATSHLLGQNFSRMFSIEFEDEKGAKQLVHQTSWGCTTRSLGVMIMTHGDDKGLVL 120
RGIQAATSHLLG NF++MF I FEDE G KQL HQTSWG TTRS+GV IM HGDD+GLV+
Sbjct 434 RGIQAATSHLLGTNFAKMFDIVFEDENGVKQLAHQTSWGFTTRSIGVSIMIHGDDQGLVI 493
Query 121 PPRVVAVQAVIIPIIFKETGGMEIVAKCRELERLLNAAGVRVKVDDRTNYTPGWKFNDWE 180
PPRV VQ VI+PII K+ +I+A ++ L AG R +DDR YTPG+KFN WE
Sbjct 494 PPRVSKVQIVIVPIIAKKELQKDIMAVAEDMYARLKKAGFRTHLDDRVGYTPGFKFNHWE 553
Query 181 LKGVPLRLEIGPRDVESCQTRVVRRDTSEARNVPWAEAASTIPAMLETMQKDLFNKAKAK 240
L+GVPLR+EIG RD+++ RV R +V +I LE +Q+ ++ +AKA+
Sbjct 554 LRGVPLRIEIGARDIQTQSCRVCYRYNGHKTDVKLETLEQSIGEALEDIQRKMYERAKAQ 613
Query 241 FEASIEQITTFDEVMPALNRRHVVLAPWCEDPETETQIKRETQRLSEIQARENADGMTGA 300
+ S+ ++ FD VMPALN + +VL PWCEDPETE++IK ETQRLS+ E +DG TG+
Sbjct 614 MDESVVKVMEFDGVMPALNSQKLVLVPWCEDPETESEIKAETQRLSQ----EGSDGKTGS 669
Query 301 MKPLCIPFDQPPMPEGTRCFFTGRPAKRWCLFGRSY 336
MK LC+P DQP MP GT+CF+TG+PA RW LFGRSY
Sbjct 670 MKCLCLPLDQPEMPPGTKCFWTGKPATRWALFGRSY 705
> cpv:cgd6_4400 proline-tRNA synthetase; class II aaRS (ybak RNA
binding domain plus tRNA synthetase) ; K01881 prolyl-tRNA
synthetase [EC:6.1.1.15]
Length=719
Score = 383 bits (984), Expect = 4e-106, Method: Compositional matrix adjust.
Identities = 198/338 (58%), Positives = 241/338 (71%), Gaps = 4/338 (1%)
Query 1 TAHATEEEAYTLVLEILELYRQWYEDYLAVPVIKGEKSENEKFAGGKKTTTIEGIIPDTG 60
TAH+T +EA +V IL Y YED LA PV+KG KSENEKF GG T +IEG IP+ G
Sbjct 384 TAHSTRKEALEMVDIILNEYASIYEDLLATPVVKGTKSENEKFPGGDITKSIEGFIPEIG 443
Query 61 RGIQAATSHLLGQNFSRMFSIEFEDEKGAKQLVHQTSWGCTTRSLGVMIMTHGDDKGLVL 120
R +QAATSHLLGQNFS+MF +EFEDEKG K+ HQTSWG TTR++GVMIMTHGD+KGLVL
Sbjct 444 RAVQAATSHLLGQNFSKMFGVEFEDEKGNKEYAHQTSWGLTTRAIGVMIMTHGDNKGLVL 503
Query 121 PPRVVAVQAVIIPIIFKETGGMEIVAKCRELERLLNAAGVRVKVDDRTNYTPGWKFNDWE 180
PP+V VQ +IIPIIFK E C E+E +L AGVRVK+DDR+NYTPGWK+N WE
Sbjct 504 PPKVAPVQVIIIPIIFKTVITEEQKKICNEVECILKKAGVRVKIDDRSNYTPGWKYNHWE 563
Query 181 LKGVPLRLEIGPRDVESCQTRVVRRDTSEARNVPWAEAASTIPAMLETMQKDLFNKAKAK 240
+KGV LR E+GPRD+E RVV RD E ++P +E S IP +LE Q L KAK +
Sbjct 564 VKGVCLRFEVGPRDIEKRSVRVVVRDNMEKMDIPISELESKIPKLLEEFQNRLLFKAKQR 623
Query 241 FEASIEQITTFDEVMPALNRRHVVLAPWCEDPETETQIKRETQRLSEIQARENADGMTGA 300
SI ++ TFD+VM LN++ +V+APWCED E +IK+ET RLS E+ MTGA
Sbjct 624 QNESIIRVDTFDKVMDTLNQKKMVIAPWCEDVSCEEEIKKETARLS--LDNEDNQSMTGA 681
Query 301 MKPLCIPFDQP-PMPEG-TRCFFTGRPAKRWCLFGRSY 336
MK LCIP DQ + EG T+CFF + AK++ LFGRSY
Sbjct 682 MKSLCIPNDQIFKIEEGKTKCFFCDKLAKKFTLFGRSY 719
> ath:AT3G62120 tRNA synthetase class II (G, H, P and S) family
protein; K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=530
Score = 376 bits (965), Expect = 8e-104, Method: Compositional matrix adjust.
Identities = 177/336 (52%), Positives = 236/336 (70%), Gaps = 12/336 (3%)
Query 1 TAHATEEEAYTLVLEILELYRQWYEDYLAVPVIKGEKSENEKFAGGKKTTTIEGIIPDTG 60
TA AT+ EA VL+ILELYR+ YE+YLAVPV+KG KSENEKFAGG TT++E IP+TG
Sbjct 207 TAFATKAEADEEVLQILELYRRIYEEYLAVPVVKGMKSENEKFAGGLYTTSVEAFIPNTG 266
Query 61 RGIQAATSHLLGQNFSRMFSIEFEDEKGAKQLVHQTSWGCTTRSLGVMIMTHGDDKGLVL 120
RG+Q ATSH LGQNF++MF I FE+EK ++V Q SW +TR++GVMIMTHGDDKGLVL
Sbjct 267 RGVQGATSHCLGQNFAKMFEINFENEKAETEMVWQNSWAYSTRTIGVMIMTHGDDKGLVL 326
Query 121 PPRVVAVQAVIIPIIFKETGGMEIVAKCRELERLLNAAGVRVKVDDRTNYTPGWKFNDWE 180
PP+V +VQ V+IP+ +K+ I C L AG+R + D R NY+PGWK++DWE
Sbjct 327 PPKVASVQVVVIPVPYKDANTQGIYDACTATASALCEAGIRAEEDLRDNYSPGWKYSDWE 386
Query 181 LKGVPLRLEIGPRDVESCQTRVVRRDTSEARNVPWAEAASTIPAMLETMQKDLFNKAKAK 240
+KGVPLR+EIGPRD+E+ Q R VRRD ++P + +LE +Q++++ AK K
Sbjct 387 MKGVPLRIEIGPRDLENDQVRTVRRDNGVKEDIPRGSLVEHVKELLEKIQQNMYEVAKQK 446
Query 241 FEASIEQITTFDEVMPALNRRHVVLAPWCEDPETETQIKRETQRLSEIQARENADGMTGA 300
EA ++++ T+DE + ALN + ++LAPWC++ E E +K T+ G TGA
Sbjct 447 REACVQEVKTWDEFIKALNEKKLILAPWCDEEEVERDVKARTK------------GETGA 494
Query 301 MKPLCIPFDQPPMPEGTRCFFTGRPAKRWCLFGRSY 336
K LC PFDQP +PEGT CF +G+PAK+W +GRSY
Sbjct 495 AKTLCSPFDQPELPEGTLCFASGKPAKKWTYWGRSY 530
> tpv:TP01_0166 prolyl-tRNA synthetase; K01881 prolyl-tRNA synthetase
[EC:6.1.1.15]
Length=683
Score = 317 bits (812), Expect = 4e-86, Method: Compositional matrix adjust.
Identities = 149/268 (55%), Positives = 196/268 (73%), Gaps = 2/268 (0%)
Query 1 TAHATEEEAYTLVLEILELYRQWYEDYLAVPVIKGEKSENEKFAGGKKTTTIEGIIPDTG 60
TAH+TE+EA V E+L+LY ++YE+YL+VPVIKG KSENEKFAG K TT++E +P G
Sbjct 359 TAHSTEKEALDTVYEMLDLYSKFYEEYLSVPVIKGVKSENEKFAGSKMTTSLEAFVPANG 418
Query 61 RGIQAATSHLLGQNFSRMFSIEFEDEKGAKQLVHQTSWGCTTRSLGVMIMTHGDDKGLVL 120
RG+QAATSHLLG FS MF I++EDE+G K+ VHQTSWG TTRS+G+MIM HGDD+GLV+
Sbjct 419 RGVQAATSHLLGTTFSDMFDIKYEDEEGEKRKVHQTSWGFTTRSIGIMIMVHGDDRGLVV 478
Query 121 PPRVVAVQAVIIPIIFKETGGMEIVAKCRELERLLNAAGVRVKVDDRTNYTPGWKFNDWE 180
PPRV Q VI+PI+ K +++++ EL +LL +G RVK+DDR YTPG+KFN WE
Sbjct 479 PPRVAKTQVVIVPILTKTAD--QVISRVNELYKLLTESGFRVKLDDRRGYTPGFKFNHWE 536
Query 181 LKGVPLRLEIGPRDVESCQTRVVRRDTSEARNVPWAEAASTIPAMLETMQKDLFNKAKAK 240
L+GVPLRLE+G +DVE+ R+VRRD +VP +TI +L+ +Q L A+ K
Sbjct 537 LRGVPLRLEVGAKDVENNTVRLVRRDNFAKADVPLENLTTTIQQLLDQIQNTLLTSAREK 596
Query 241 FEASIEQITTFDEVMPALNRRHVVLAPW 268
E SI + F+EVM ALN +++APW
Sbjct 597 MEQSIVKAYDFEEVMVALNNDKLIMAPW 624
> sce:YHR020W Protein of unknown function that may interact with
ribosomes, based on co-purification experiments; has similarity
to proline-tRNA ligase; YHR020W is an essential gene
(EC:6.1.1.15); K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=688
Score = 313 bits (802), Expect = 7e-85, Method: Compositional matrix adjust.
Identities = 164/344 (47%), Positives = 220/344 (63%), Gaps = 8/344 (2%)
Query 1 TAHATEEEAYTLVLEILELYRQWYEDYLAVPVIKGEKSENEKFAGGKKTTTIEGIIPDTG 60
TAH T ++A VL+IL+ Y YE+ LAVPV+KG K+E EKFAGG TTT EG IP TG
Sbjct 345 TAHLTAKDAEEEVLQILDFYAGVYEELLAVPVVKGRKTEKEKFAGGDFTTTCEGYIPQTG 404
Query 61 RGIQAATSHLLGQNFSRMFSIEFEDEKGA---KQLVHQTSWGCTTRSLGVMIMTHGDDKG 117
RGIQ ATSH LGQNFS+MF++ E+ G+ K +Q SWG +TR +GVM+M H D+KG
Sbjct 405 RGIQGATSHHLGQNFSKMFNLSVENPLGSDHPKIFAYQNSWGLSTRVIGVMVMIHSDNKG 464
Query 118 LVLPPRVVAVQAVIIPI-IFKETGGME---IVAKCRELERLLNAAGVRVKVDDRTNYTPG 173
LV+PPRV Q+V+IP+ I K+T + I R +E L G+R D NYTPG
Sbjct 465 LVIPPRVSQFQSVVIPVGITKKTSEEQRKHIHETARSVESRLKKVGIRAFGDYNDNYTPG 524
Query 174 WKFNDWELKGVPLRLEIGPRDVESCQTRVVRRDTSEARNVPWAEAASTIPAMLETMQKDL 233
WKF+ +ELKG+P+R+E+GP+D+E Q VVRR+ S+ V + E + IP +LE MQ DL
Sbjct 525 WKFSQYELKGIPIRIELGPKDIEKNQVVVVRRNDSKKYVVSFDELEARIPEILEEMQGDL 584
Query 234 FNKAKAKFEASIEQITTFDEVMPALNRRHVVLAPWCEDPETETQIKRET-QRLSEIQARE 292
F KAK F+ + + +PALN+++V+LAPWC E E IK + ++ + E
Sbjct 585 FKKAKELFDTHRVIVNEWSGFVPALNKKNVILAPWCGVMECEEDIKESSAKKDDGEEFEE 644
Query 293 NADGMTGAMKPLCIPFDQPPMPEGTRCFFTGRPAKRWCLFGRSY 336
+ + K LCIPFDQP + EG +C R A +C+FGRSY
Sbjct 645 DDKAPSMGAKSLCIPFDQPVLNEGQKCIKCERIAVNYCMFGRSY 688
> hsa:2058 EPRS, DKFZp313B047, EARS, GLUPRORS, PARS, QARS, QPRS;
glutamyl-prolyl-tRNA synthetase (EC:6.1.1.15 6.1.1.17); K14163
bifunctional glutamyl/prolyl-tRNA synthetase [EC:6.1.1.17
6.1.1.15]
Length=1512
Score = 310 bits (794), Expect = 5e-84, Method: Compositional matrix adjust.
Identities = 167/345 (48%), Positives = 213/345 (61%), Gaps = 15/345 (4%)
Query 1 TAHATEEEAYTLVLEILELYRQWYEDYLAVPVIKGEKSENEKFAGGKKTTTIEGIIPDTG 60
+A AT EEA VL+IL+LY Q YE+ LA+PV+KG K+E EKFAGG TTTIE I +G
Sbjct 1174 SAFATMEEAAEEVLQILDLYAQVYEELLAIPVVKGRKTEKEKFAGGDYTTTIEAFISASG 1233
Query 61 RGIQAATSHLLGQNFSRMFSIEFEDEK--GAKQLVHQTSWGCTTRSLGVMIMTHGDDKGL 118
R IQ TSH LGQNFS+MF I FED K G KQ +Q SWG TTR++GVM M HGD+ GL
Sbjct 1234 RAIQGGTSHHLGQNFSKMFEIVFEDPKIPGEKQFAYQNSWGLTTRTIGVMTMVHGDNMGL 1293
Query 119 VLPPRVVAVQAVIIPI----IFKETGGMEIVAKCRELERLLNAAGVRVKVDDRTNYTPGW 174
VLPPRV VQ VIIP E ++AKC + R L + +RV+ D R NY+PGW
Sbjct 1294 VLPPRVACVQVVIIPCGITNALSEEDKEALIAKCNDYRRRLLSVNIRVRADLRDNYSPGW 1353
Query 175 KFNDWELKGVPLRLEIGPRDVESCQTRVVRRDTSEARNVPWAEAASTIPAMLETMQKDLF 234
KFN WELKGVP+RLE+GPRD++SCQ VRRDT E V EA + + A+LE +Q LF
Sbjct 1354 KFNHWELKGVPIRLEVGPRDMKSCQFVAVRRDTGEKLTVAENEAETKLQAILEDIQVTLF 1413
Query 235 NKAKAKFEASIEQITTFDEVMPALNRRHVVLAPWCEDPETETQIKRETQRLSEIQARENA 294
+A + + T ++ L+ +V P+C + + E IK+ T R +++ +
Sbjct 1414 TRASEDLKTHMVVANTMEDFQKILDSGKIVQIPFCGEIDCEDWIKKTTARDQDLEPGAPS 1473
Query 295 DGMTGAMKPLCIPFDQPPMPE---GTRCFFTGRPAKRWCLFGRSY 336
G K LCIPF P+ E G +C PAK + LFGRSY
Sbjct 1474 MG----AKSLCIPFK--PLCELQPGAKCVCGKNPAKYYTLFGRSY 1512
> mmu:107508 Eprs, 2410081F06Rik, 3010002K18Rik, C79379, Qprs;
glutamyl-prolyl-tRNA synthetase (EC:6.1.1.15 6.1.1.17); K14163
bifunctional glutamyl/prolyl-tRNA synthetase [EC:6.1.1.17
6.1.1.15]
Length=1512
Score = 306 bits (785), Expect = 6e-83, Method: Compositional matrix adjust.
Identities = 166/345 (48%), Positives = 215/345 (62%), Gaps = 15/345 (4%)
Query 1 TAHATEEEAYTLVLEILELYRQWYEDYLAVPVIKGEKSENEKFAGGKKTTTIEGIIPDTG 60
+A AT EEA VL+ILELY + YE+ LA+PV++G K+E EKFAGG TTTIE I +G
Sbjct 1174 SAFATFEEAADEVLQILELYARVYEELLAIPVVRGRKTEKEKFAGGDYTTTIEAFISASG 1233
Query 61 RGIQAATSHLLGQNFSRMFSIEFEDEK--GAKQLVHQTSWGCTTRSLGVMIMTHGDDKGL 118
R IQ ATSH LGQNFS+M I FED K G KQ +Q SWG TTR++GVM+M HGD+ GL
Sbjct 1234 RAIQGATSHHLGQNFSKMCEIVFEDPKTPGEKQFAYQCSWGLTTRTIGVMVMVHGDNMGL 1293
Query 119 VLPPRVVAVQAVIIPI----IFKETGGMEIVAKCRELERLLNAAGVRVKVDDRTNYTPGW 174
VLPPRV +VQ V+IP E ++AKC E R L A +RV+VD R NY+PGW
Sbjct 1294 VLPPRVASVQVVVIPCGITNALSEEDREALMAKCNEYRRRLLGANIRVRVDLRDNYSPGW 1353
Query 175 KFNDWELKGVPLRLEIGPRDVESCQTRVVRRDTSEARNVPWAEAASTIPAMLETMQKDLF 234
KFN WELKGVP+RLE+GPRD++SCQ VRRDT E + EA + + +LE +Q +LF
Sbjct 1354 KFNHWELKGVPVRLEVGPRDMKSCQFVAVRRDTGEKLTIAEKEAEAKLEKVLEDIQLNLF 1413
Query 235 NKAKAKFEASIEQITTFDEVMPALNRRHVVLAPWCEDPETETQIKRETQRLSEIQARENA 294
+A + + T ++ L+ V P+C + + E IK+ T R +++ +
Sbjct 1414 TRASEDLKTHMVVSNTLEDFQKVLDAGKVAQIPFCGEIDCEDWIKKMTARDQDVEPGAPS 1473
Query 295 DGMTGAMKPLCIPFDQPPMPE---GTRCFFTGRPAKRWCLFGRSY 336
G K LCIPF+ P+ E G C PAK + LFGRSY
Sbjct 1474 MG----AKSLCIPFN--PLCELQPGAMCVCGKNPAKFYTLFGRSY 1512
> dre:562037 eprs, wu:fb38d08, wu:ft34d10; glutamyl-prolyl-tRNA
synthetase; K14163 bifunctional glutamyl/prolyl-tRNA synthetase
[EC:6.1.1.17 6.1.1.15]
Length=1511
Score = 294 bits (752), Expect = 4e-79, Method: Compositional matrix adjust.
Identities = 158/343 (46%), Positives = 208/343 (60%), Gaps = 11/343 (3%)
Query 1 TAHATEEEAYTLVLEILELYRQWYEDYLAVPVIKGEKSENEKFAGGKKTTTIEGIIPDTG 60
TA AT+EEA VL+IL+LY + YE+ +A+PV+KG K+E EKFAGG TTT+E I +G
Sbjct 1173 TAFATKEEATEEVLQILDLYARVYEELMAIPVVKGRKTEKEKFAGGDYTTTVEAFISASG 1232
Query 61 RGIQAATSHLLGQNFSRMFSIEFEDEK--GAKQLVHQTSWGCTTRSLGVMIMTHGDDKGL 118
R IQ ATSH LGQNFSRMF I FED K G KQL +Q SWG TTR++GV+ M HGD+ GL
Sbjct 1233 RAIQGATSHHLGQNFSRMFEIVFEDPKKPGEKQLAYQNSWGITTRTIGVLTMVHGDNMGL 1292
Query 119 VLPPRVVAVQAVIIPI----IFKETGGMEIVAKCRELERLLNAAGVRVKVDDRTNYTPGW 174
VLPPRV +Q +IIP E ++ +C + L A +RVK D R NY+PGW
Sbjct 1293 VLPPRVACLQVIIIPCGITATLPEAEKDLLLDQCSKYLSRLQKADIRVKADLRDNYSPGW 1352
Query 175 KFNDWELKGVPLRLEIGPRDVESCQTRVVRRDTSEARNVPWAEAASTIPAMLETMQKDLF 234
KFN WELKGVP+R E+GP+D++ Q VRRDT E V A+A IP +LE +Q +LF
Sbjct 1353 KFNHWELKGVPIRFEVGPKDLKQGQFVAVRRDTGEKITVKEADAEKKIPNLLEEIQNNLF 1412
Query 235 NKAKAKFEASIEQITTFDEVMPALNRRHVVLAPWCEDPETETQIKRETQRLSEIQARENA 294
+A + T ++ L++ +V P+C E E IK+ T + +++ +
Sbjct 1413 KRASDDLNKHMVVADTMEQFQKDLDQGRIVQIPFCGGIECEDWIKKTTAKDQDLEPGAPS 1472
Query 295 DGMTGAMKPLCIPFDQ-PPMPEGTRCFFTGRPAKRWCLFGRSY 336
G K LCIPF + G C PA+ + LFGRSY
Sbjct 1473 MG----AKSLCIPFKPLKNLQAGQMCVSGKEPAQYYTLFGRSY 1511
> xla:100158287 hypothetical protein LOC100158287; K01881 prolyl-tRNA
synthetase [EC:6.1.1.15]
Length=655
Score = 292 bits (748), Expect = 9e-79, Method: Compositional matrix adjust.
Identities = 159/346 (45%), Positives = 214/346 (61%), Gaps = 17/346 (4%)
Query 1 TAHATEEEAYTLVLEILELYRQWYEDYLAVPVIKGEKSENEKFAGGKKTTTIEGIIPDTG 60
TA AT EEA VL+IL+LY + YED LA+PV+KG K+E EKFAGG TTT+E I +
Sbjct 317 TAFATYEEAAEEVLQILDLYARVYEDLLAIPVVKGRKTEKEKFAGGDYTTTVEAFISASS 376
Query 61 RGIQAATSHLLGQNFSRMFSIEFEDEK--GAKQLVHQTSWGCTTRSLGVMIMTHGDDKGL 118
R IQ ATSH LGQNFS++F I FED K G KQ +Q SWG +TR++G M+M H D+ G+
Sbjct 377 RAIQGATSHHLGQNFSKIFEIVFEDPKTPGLKQYAYQNSWGLSTRTIGAMVMVHADNIGM 436
Query 119 VLPPRVVAVQAVIIPIIFKETGGME----IVAKCRE-LERLLNAAGVRVKVDDRTNYTPG 173
VLPPRV VQ VIIP T E ++ KC + L+RLLNA VR + D R NY+PG
Sbjct 437 VLPPRVACVQVVIIPCGITNTLSEEDKDALLKKCDQYLKRLLNA-DVRARADLRDNYSPG 495
Query 174 WKFNDWELKGVPLRLEIGPRDVESCQTRVVRRDTSEARNVPWAEAASTIPAMLETMQKDL 233
WKFN WELKGVP+RLE+GPRD+++ Q VRRD E + +P +A + + +LE + +L
Sbjct 496 WKFNHWELKGVPIRLEVGPRDMKNKQFVAVRRDNGEKQTIPEGQAETRLKEILEEIHTNL 555
Query 234 FNKAKAKFEASIEQITTFDEVMPALNRRHVVLAPWCEDPETETQIKRETQRLSEIQAREN 293
+N+A + + + ++ L + +V P+C + E E IK+ T R +++
Sbjct 556 YNRAHSDLTSHMVVANNMEDFQKHLEKGRIVQIPFCGEIECEDWIKKTTARDQDLEPGAP 615
Query 294 ADGMTGAMKPLCIPFDQPPMPE---GTRCFFTGRPAKRWCLFGRSY 336
+ G K LCIPF P+ E G +C AK + LFGRSY
Sbjct 616 SMG----AKSLCIPFK--PLSELQPGAKCVCEKNAAKFYTLFGRSY 655
> xla:100158438 eprs; glutamyl-prolyl-tRNA synthetase; K14163
bifunctional glutamyl/prolyl-tRNA synthetase [EC:6.1.1.17 6.1.1.15]
Length=1499
Score = 289 bits (739), Expect = 1e-77, Method: Compositional matrix adjust.
Identities = 158/346 (45%), Positives = 212/346 (61%), Gaps = 17/346 (4%)
Query 1 TAHATEEEAYTLVLEILELYRQWYEDYLAVPVIKGEKSENEKFAGGKKTTTIEGIIPDTG 60
TA AT EEA VL+IL+LY + YE+ LA+PV+KG K+E EKFAGG TTT+E I +
Sbjct 1161 TAFATYEEAAEEVLQILDLYARVYEELLAIPVVKGRKTEKEKFAGGDYTTTVEAFISASS 1220
Query 61 RGIQAATSHLLGQNFSRMFSIEFEDEK--GAKQLVHQTSWGCTTRSLGVMIMTHGDDKGL 118
R IQ ATSH LGQNFS+MF I FED K G KQ +Q SWG +TR++G M+M H D+ G+
Sbjct 1221 RAIQGATSHHLGQNFSKMFEIVFEDPKTPGLKQYAYQNSWGLSTRTIGAMVMVHADNTGM 1280
Query 119 VLPPRVVAVQAVIIPIIFKETGGME----IVAKCRE-LERLLNAAGVRVKVDDRTNYTPG 173
VLPPRV VQ +IIP T E ++ KC + L+RLLN VR + D R NY+PG
Sbjct 1281 VLPPRVACVQIIIIPCGITNTLAEEDKDALLKKCDQYLKRLLNV-DVRARADLRDNYSPG 1339
Query 174 WKFNDWELKGVPLRLEIGPRDVESCQTRVVRRDTSEARNVPWAEAASTIPAMLETMQKDL 233
WKFN WELKGVP+RLE+GPRD+++ Q VRRD E + +P +A + + +LE + +L
Sbjct 1340 WKFNHWELKGVPIRLEVGPRDMKNKQFVAVRRDNGEKQTIPEDQAETRLKEILEEIHTNL 1399
Query 234 FNKAKAKFEASIEQITTFDEVMPALNRRHVVLAPWCEDPETETQIKRETQRLSEIQAREN 293
+N+A + + +E L + +V P+C + E E IK+ T R +++
Sbjct 1400 YNRALGDLTSHMVVANNMEEFQKHLEKGQIVQIPFCGEIECEDWIKKITARDQDLEPGAP 1459
Query 294 ADGMTGAMKPLCIPFDQPPMPE---GTRCFFTGRPAKRWCLFGRSY 336
+ G K LCIPF P+ E G +C AK + LFGRSY
Sbjct 1460 SMG----AKSLCIPFK--PLSELQPGAKCVCEKNAAKFYTLFGRSY 1499
> cel:T20H4.3 prs-1; Prolyl tRNA Synthetase family member (prs-1);
K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=581
Score = 288 bits (736), Expect = 3e-77, Method: Compositional matrix adjust.
Identities = 144/340 (42%), Positives = 204/340 (60%), Gaps = 11/340 (3%)
Query 1 TAHATEEEAYTLVLEILELYRQWYEDYLAVPVIKGEKSENEKFAGGKKTTTIEGIIPDTG 60
TA A +A V +IL+LY Y D LA+PV+KG KSE EKFAGG TTT+E + G
Sbjct 249 TAFANPADAEKEVFQILDLYAGVYTDLLAIPVVKGRKSEKEKFAGGDFTTTVEAYVACNG 308
Query 61 RGIQAATSHLLGQNFSRMFSIEFED--EKGAKQLVHQTSWGCTTRSLGVMIMTHGDDKGL 118
RGIQ ATSH LGQNFS+MF I +ED ++G + Q SWG +TR++G M+M HGDDKGL
Sbjct 309 RGIQGATSHHLGQNFSKMFDISYEDPAKEGERAFAWQNSWGLSTRTIGAMVMIHGDDKGL 368
Query 119 VLPPRVVAVQAVIIPIIFKETGGMEIVAKCRELERLLNAAGVRVKVDDRTNYTPGWKFND 178
VLPPRV AVQ +++P+ FK + ++ + L A GV+ + D R NY GWKFN
Sbjct 369 VLPPRVAAVQVIVVPVGFKTENKQSLFDAVDKVTKDLVADGVKAEHDLRENYNAGWKFNH 428
Query 179 WELKGVPLRLEIGPRDVESCQTRVVRRDTSEARNVPWAEAASTIPAMLETMQKDLFNKAK 238
WELKGVP+R E+GP D+ Q V R E + + + T+ ++L+ + ++NK
Sbjct 429 WELKGVPIRFEVGPNDLAKNQVTAVIRHNGEKKQLSLEGLSKTVKSLLDEIHTAMYNKVL 488
Query 239 AKFEASIEQITTFDEVMPALNRRHVVLAPWCEDPETETQIKRETQRLSEIQARENADGMT 298
A+ ++ ++ T +E L+ + ++L+P+C P+ E +IK+ + RE+ +G
Sbjct 489 AQRDSHLKSTTCIEEFKKLLDEKCIILSPFCGRPDCEEEIKKAS-------TREDGEGAQ 541
Query 299 GAMKPLCIPFDQPPMPEGTRCFFTG--RPAKRWCLFGRSY 336
K LCIP DQP ++C F AK + LFGRSY
Sbjct 542 MGAKTLCIPLDQPKQELPSKCLFPSCTYNAKAFALFGRSY 581
> ath:AT5G52520 OVA6; OVA6 (OVULE ABORTION 6); ATP binding / aminoacyl-tRNA
ligase/ nucleotide binding / proline-tRNA ligase;
K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=543
Score = 233 bits (595), Expect = 6e-61, Method: Compositional matrix adjust.
Identities = 139/343 (40%), Positives = 195/343 (56%), Gaps = 28/343 (8%)
Query 1 TAHATEEEAYTLVLEILELYRQWYEDYLAVPVIKGEKSENEKFAGGKKTTTIEGIIPDTG 60
TAHAT EEA +++E+Y ++ + A+PVI G KS+ E FAG T TIE ++ D
Sbjct 222 TAHATPEEAEKEAKQMIEIYTRFAFEQTAIPVIPGRKSKLETFAGADITYTIEAMMGDR- 280
Query 61 RGIQAATSHLLGQNFSRMFSIEFEDEKGAKQLVHQTSWGCTTRSLGVMIMTHGDDKGLVL 120
+ +QA TSH LGQNFSR F +F DE G +Q V QTSW +TR +G +IMTHGDD GL+L
Sbjct 281 KALQAGTSHNLGQNFSRAFGTQFADENGERQHVWQTSWAVSTRFVGGIIMTHGDDTGLML 340
Query 121 PPRVVAVQAVIIPIIFKETGGMEIVAKCRELERLLNAAGVRVKVDDRTNYTPGWKFNDWE 180
PP++ +Q VI+PI K+T +++ ++ L AGVRVK+DD TPGWKFN WE
Sbjct 341 PPKIAPIQVVIVPIWKKDTEKTGVLSAASSVKEALQTAGVRVKLDDTDQRTPGWKFNFWE 400
Query 181 LKGVPLRLEIGPRDVESCQTRVVRRDT--SEARNVPWAEAASTIPAM----LETMQKDLF 234
+KG+PLR+EIGPRDV S V RRD + + ST+ A L+ +Q L
Sbjct 401 MKGIPLRIEIGPRDVSSNSVVVSRRDVPGKAGKVFGISMEPSTLVAYVKEKLDEIQTSLL 460
Query 235 NKAKAKFEASIEQITTFDEVMPALNRRHVVLAPWCEDPETETQIKRETQRLSEIQARENA 294
KA + +++I + ++ E+ A++ PW E ++K E
Sbjct 461 EKALSFRDSNIVDVNSYAELKDAISSGKWARGPWSASDADEQRVKEE------------- 507
Query 295 DGMTGAMKPLCIPFDQPPMPEGTR-CFFTGRPAKRWCLFGRSY 336
TGA C PF+Q +GT+ C TG PA+ +F +SY
Sbjct 508 ---TGATI-RCFPFEQ---TQGTKTCLMTGNPAEEVAIFAKSY 543
> ath:AT5G10880 tRNA synthetase-related / tRNA ligase-related
Length=309
Score = 188 bits (477), Expect = 3e-47, Method: Compositional matrix adjust.
Identities = 95/228 (41%), Positives = 136/228 (59%), Gaps = 8/228 (3%)
Query 110 MTHGDDKGLVLPPRVVAVQAVIIPIIFKETGGM-EIVAKCRELERLLNAAGVRVKVDDRT 168
MTHGDDKGLV PP+V VQ V+I + K E+ C +E L AG+R + D R
Sbjct 89 MTHGDDKGLVFPPKVAPVQVVVIHVPIKGAADYQELCDACEAVESTLLGAGIRAEADIRD 148
Query 169 NYTPGWKFNDWELKGVPLRLEIGPRDVESCQTRVVRRDTSEARNVPWAEAASTIPAMLET 228
NY+ GWK+ D EL GVPLR+E GPRD+ + Q R+V RD +V + + +LE
Sbjct 149 NYSCGWKYADQELTGVPLRIETGPRDLANDQVRIVTRDNGAKMDVKRGDLIEQVKDLLEK 208
Query 229 MQKDLFNKAKAKFEASIEQITTFDEVMPALNRRHVVLAPWCEDPETETQIKRETQRLSEI 288
+Q +L++ AK K E +++ T+DE + AL+++ ++LAPWC+ E E +KR T+
Sbjct 209 IQSNLYDVAKRKVEECTQKVETWDEFVEALSQKKLILAPWCDKVEVEKDVKRRTR----- 263
Query 289 QARENADGMTGAMKPLCIPFDQPPMPEGTRCFFTGRPAKRWCLFGRSY 336
+ G G K LC P +QP + E T CF +G+PAK+W +GRSY
Sbjct 264 --GDETGGGGGGAKTLCTPLEQPELGEETLCFASGKPAKKWSYWGRSY 309
> eco:b0194 proS, drpA, ECK0194, JW0190; prolyl-tRNA synthetase
(EC:6.1.1.15); K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=572
Score = 60.1 bits (144), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 39/138 (28%), Positives = 69/138 (50%), Gaps = 4/138 (2%)
Query 61 RGIQAATSHLLGQNFSRMFSIEFEDEKGAKQLVHQTSWGC-TTRSLGVMIMTHGDDKGLV 119
RGI+ LG +S + E G Q++ +G TR + I + D++G+V
Sbjct 407 RGIEVGHIFQLGTKYSEALKASVQGEDGRNQILTMGCYGIGVTRVVAAAIEQNYDERGIV 466
Query 120 LPPRVVAVQAVIIPIIFKETGGMEIVAKCRELERLLNAAGVRVKVDDRTNYTPGWKFNDW 179
P + Q I+P+ ++ ++ +A+ +L L A G+ V +DDR PG F D
Sbjct 467 WPDAIAPFQVAILPMNMHKSFRVQELAE--KLYSELRAQGIEVLLDDRKE-RPGVMFADM 523
Query 180 ELKGVPLRLEIGPRDVES 197
EL G+P + +G R++++
Sbjct 524 ELIGIPHTIVLGDRNLDN 541
> ath:AT2G04842 EMB2761 (EMBRYO DEFECTIVE 2761); ATP binding /
aminoacyl-tRNA ligase/ ligase, forming aminoacyl-tRNA and related
compounds / nucleotide binding / threonine-tRNA ligase
(EC:6.1.1.3); K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=650
Score = 47.0 bits (110), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 48/189 (25%), Positives = 80/189 (42%), Gaps = 19/189 (10%)
Query 43 FAGGKKTTTIEGIIPDTGRGIQAATSHLLGQNFSRMFSIEFEDEKGAKQ---LVHQTSWG 99
F G K IE + GR Q +T + N + F I + D K+ ++H+ G
Sbjct 474 FYGPKIDLKIEDAL---GRKWQCSTIQV-DFNLPQRFDITYVDTNSDKKRPIMIHRAVLG 529
Query 100 CTTRSLGVMIMTHGDDKGLVLPPRVVAVQAVIIPIIFKETGGMEIVAKCRELERLLNAAG 159
R GV+I + D L L P VQ ++P+ + + C+E+ + L A G
Sbjct 530 SLERFFGVLIEHYAGDFPLWLSP----VQVRVLPVTDNQ------LEFCKEVSKKLRACG 579
Query 160 VRVKVDDRTNYTPGWKFNDWELKGVPLRLEIGPRDVESCQTRVVRRDTSEARNVPWAEAA 219
VR ++ + E + +PL +GP++VE+ V R E +P +
Sbjct 580 VRAELCHGERLPK--LIRNAETQKIPLMAVVGPKEVETGTVTVRSRFGGELGTIPVDDLI 637
Query 220 STIPAMLET 228
+ I +ET
Sbjct 638 NKINIAVET 646
> mmu:230577 Pars2, BC027073, MGC47008; prolyl-tRNA synthetase
(mitochondrial)(putative) (EC:6.1.1.15); K01881 prolyl-tRNA
synthetase [EC:6.1.1.15]
Length=511
Score = 43.1 bits (100), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 39/141 (27%), Positives = 59/141 (41%), Gaps = 6/141 (4%)
Query 61 RGIQAATSHLLGQNFSRMFSIEFEDEKGAKQLVHQTSWGC-TTRSLGVMIMTHGDDKGLV 119
+GI+ + L +S +F+ F + G L +G TR L I + +
Sbjct 339 KGIEVGHTFYLSTKYSSIFNALFTNAHGESLLAEMGCYGLGVTRILAAAIEVLSTEDCIR 398
Query 120 LPPRVVAVQAVIIPII--FKETGGMEIVAKCRE--LERLLNAAGVRVKVDDRTNYTPGWK 175
P + Q +IP KET EIV K + +E + G V +DDRT+ T G +
Sbjct 399 WPRLLAPYQVCVIPPKKGSKETAATEIVEKLYDDIMEAVPQLRG-EVLLDDRTHLTIGNR 457
Query 176 FNDWELKGVPLRLEIGPRDVE 196
D G P + G R +E
Sbjct 458 LKDANKLGYPFVIIAGKRALE 478
> hsa:25973 PARS2, DKFZp727A071, MGC14416, MGC19467, MT-PRORS;
prolyl-tRNA synthetase 2, mitochondrial (putative) (EC:6.1.1.15);
K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=475
Score = 42.4 bits (98), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 35/140 (25%), Positives = 59/140 (42%), Gaps = 4/140 (2%)
Query 61 RGIQAATSHLLGQNFSRMFSIEFEDEKGAKQLVHQTSWGC-TTRSLGVMIMTHGDDKGLV 119
+GI+ + LG +S +F+ +F + G L +G TR L I + +
Sbjct 303 KGIEVGHTFYLGTKYSSIFNAQFTNVCGKPTLAEMGCYGLGVTRILAAAIEVLSTEDCVR 362
Query 120 LPPRVVAVQAVIIPII--FKETGGMEIVAKCRE-LERLLNAAGVRVKVDDRTNYTPGWKF 176
P + QA +IP KE E++ + + + + V +DDRT+ T G +
Sbjct 363 WPSLLAPYQACLIPPKKGSKEQAASELIGQLYDHITEAVPQLHGEVLLDDRTHLTIGNRL 422
Query 177 NDWELKGVPLRLEIGPRDVE 196
D G P + G R +E
Sbjct 423 KDANKFGYPFVIIAGKRALE 442
> eco:b1719 thrS, ECK1717, JW1709; threonyl-tRNA synthetase (EC:6.1.1.3);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=642
Score = 42.4 bits (98), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 37/159 (23%), Positives = 74/159 (46%), Gaps = 18/159 (11%)
Query 84 EDEKGAKQLVHQTSWGCTTRSLGVMIMTHGDDKGLVLPPRVVAVQAVIIPIIFKETGGME 143
++E+ ++H+ G R +G++ ++ P + VQ VI+ I ++
Sbjct 501 DNERKVPVMIHRAILGSMERFIGILT----EEFAGFFPTWLAPVQVVIMNITDSQS---- 552
Query 144 IVAKCRELERLLNAAGVRVKVDDRTNYTPGWKFNDWELKGVPLRLEIGPRDVESCQTRVV 203
EL + L+ AG+RVK D R N G+K + L+ VP L G ++VES + V
Sbjct 553 --EYVNELTQKLSNAGIRVKADLR-NEKIGFKIREHTLRRVPYMLVCGDKEVESGKVAVR 609
Query 204 RRDTSEARNVPWAEAASTIPAMLETMQKDLFNKAKAKFE 242
R + + + ++E +Q+++ +++ + E
Sbjct 610 TRRGKDL-------GSMDVNEVIEKLQQEIRSRSLKQLE 641
> dre:325494 pars2, fc83e06, wu:fc83e06, zgc:77163; prolyl-tRNA
synthetase (mitochondrial) (EC:6.1.1.15); K01881 prolyl-tRNA
synthetase [EC:6.1.1.15]
Length=477
Score = 39.7 bits (91), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 35/137 (25%), Positives = 55/137 (40%), Gaps = 8/137 (5%)
Query 61 RGIQAATSHLLGQNFSRMFSIEFEDEKGAKQLVHQTSWGC-TTRSLGVMIMTHGDDKGLV 119
RGI+ + LG +S+ F F + + +G TR L I + +
Sbjct 305 RGIEVGHTFYLGTKYSQAFKALFVNASNVPAVAEMGCFGLGVTRILAASIEVMSTEDAIH 364
Query 120 LPPRVVAVQAVIIPIIFKETGGMEIVAKCRELERLL-----NAAGVRVKVDDRTNYTPGW 174
P + Q ++P K + E K EL + L N G V +DDRT T G
Sbjct 365 WPGLIAPYQVCVLPPK-KGSKAHEATQKAEELAQSLGNSLPNLRG-EVVLDDRTQMTIGK 422
Query 175 KFNDWELKGVPLRLEIG 191
+ D ++ G P + +G
Sbjct 423 RLKDAKILGYPFVVVVG 439
> tpv:TP02_0040 prolyl-tRNA synthetase; K01881 prolyl-tRNA synthetase
[EC:6.1.1.15]
Length=461
Score = 37.4 bits (85), Expect = 0.072, Method: Compositional matrix adjust.
Identities = 37/137 (27%), Positives = 59/137 (43%), Gaps = 12/137 (8%)
Query 58 DTGRGIQAATSHLL--GQNFSRMFSIEFEDEKGAKQLVHQTSWGC-TTRSLGVMIMTHGD 114
D + HL G ++S ++ ++FE ++ ++ S+G R L V+I + D
Sbjct 257 DQNSNTELEVGHLFKFGTHYSDVYGLKFEISGRKREKLYMNSYGIGINRLLNVLINNYSD 316
Query 115 DKGLVLPPRVVAVQAVIIPIIFKETGGMEIVAKCRELERLLNAAGVRVKVDDRTNYTPGW 174
+ G+ LP + II T G E+ R L G+ V DDR+ T
Sbjct 317 EFGIKLPQLIAPYDVTIID---STTNG-----DGWEITRGLMKKGLTVLYDDRSE-TIKN 367
Query 175 KFNDWELKGVPLRLEIG 191
K D + GVP L +G
Sbjct 368 KLLDSKRIGVPHILILG 384
> hsa:80222 TARS2, FLJ12528, TARSL1; threonyl-tRNA synthetase
2, mitochondrial (putative) (EC:6.1.1.3); K01868 threonyl-tRNA
synthetase [EC:6.1.1.3]
Length=718
Score = 36.2 bits (82), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 38/151 (25%), Positives = 63/151 (41%), Gaps = 14/151 (9%)
Query 60 GRGIQAATSHLLGQNFSRMFSIEFEDEKGAKQ---LVHQTSWGCTTRSLGVMIMTHGDDK 116
GR Q T L Q R F ++++ + GA + L+H+ G R LGV+ + G
Sbjct 549 GRPHQCGTIQLDFQLPLR-FDLQYKGQAGALERPVLIHRAVLGSVERLLGVLAESCGGKW 607
Query 117 GLVLPPRVVAVQAVIIPIIFKETGGMEIVAKCRELERLLNAAGVRVKVDDRTNYTPGWKF 176
L L P Q V+IP+ G E +E ++ L AAG+ +D + T +
Sbjct 608 PLWLSP----FQVVVIPV------GSEQEEYAKEAQQSLRAAGLVSDLDADSGLTLSRRI 657
Query 177 NDWELKGVPLRLEIGPRDVESCQTRVVRRDT 207
+L + +G ++ + RD
Sbjct 658 RRAQLAHYNFQFVVGQKEQSKRTVNIRTRDN 688
> pfa:PFI1240c prolyl-t-RNA synthase, putative (EC:6.1.1.15);
K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=579
Score = 35.8 bits (81), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 30/121 (24%), Positives = 54/121 (44%), Gaps = 14/121 (11%)
Query 18 ELYRQWYEDY-LAVPVIKGEKSENEKFAGGKKTTTIEG----IIPDTGRGIQAATSHLLG 72
E Y+ +E+ L+ +IK K + K +E ++ G+ +AA LG
Sbjct 256 ECYKNIFEELKLSFNIIKKRKKD--------KMNALESHEFQVLSRDGKYKEAAHIFKLG 307
Query 73 QNFSRMFSIEFEDEKGAKQLVHQTSWGC-TTRSLGVMIMTHGDDKGLVLPPRVVAVQAVI 131
+S I++ D+K K+ + S+G R L +I D++G+ LP +V +
Sbjct 308 DYYSNKLDIKYLDKKNEKKNILMGSYGIGIYRLLYFLIENFYDEEGIKLPQQVAPFSVYL 367
Query 132 I 132
I
Sbjct 368 I 368
> mmu:71807 Tars2, 2610024N01Rik, AI429208, Tarsl1; threonyl-tRNA
synthetase 2, mitochondrial (putative) (EC:6.1.1.3); K01868
threonyl-tRNA synthetase [EC:6.1.1.3]
Length=697
Score = 35.4 bits (80), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 42/171 (24%), Positives = 72/171 (42%), Gaps = 17/171 (9%)
Query 60 GRGIQAATSHLLGQNFSRMFSIEFEDEKGAKQ---LVHQTSWGCTTRSLGVMIMTHGDDK 116
GR Q T L Q R F ++++ G + L+H+ G R LGV+ + G
Sbjct 528 GRPHQCGTIQLDFQLPLR-FDLQYKGPAGTPECPVLIHRAVLGSVERLLGVLAESCGGKW 586
Query 117 GLVLPPRVVAVQAVIIPIIFKETGGMEIVAKCRELERLLNAAGVRVKVDDRTNYTPGWKF 176
L L P +Q V+IP+ E R++++ L AAG+ +D + T +
Sbjct 587 PLWLSP----LQVVVIPV------RTEQEEYARQVQQCLQAAGLVSDLDADSGLTLSRRV 636
Query 177 NDWELKGVPLRLEIGPRDVESCQTRVVRRDTSEARNVPWAEAASTIPAMLE 227
+L + +G R+ R V T + R + + A ++ +LE
Sbjct 637 RRAQLAHYNFQFVVGQREQSQ---RTVNVRTRDNRQLGERDLAESVQRLLE 684
> dre:558958 lamc3, si:dkey-245l10.1; laminin, gamma 3; K06247
laminin, gamma 3
Length=1643
Score = 33.9 bits (76), Expect = 0.94, Method: Composition-based stats.
Identities = 20/65 (30%), Positives = 35/65 (53%), Gaps = 3/65 (4%)
Query 230 QKDLFNKAKAKFEASIEQITTFDEVMPALNRRHVVLAPWCEDPETETQIKRETQRLSEIQ 289
+ ++ K+ A IEQ+ ++V NR ++ L ED TE+ I+ T++L E+Q
Sbjct 1193 ESEVLVKSHTDMAAHIEQVA--EDVFKMANRTYLQLVTLLEDNSTESHIQDLTEKLEEMQ 1250
Query 290 A-REN 293
+EN
Sbjct 1251 QLKEN 1255
> xla:379241 pars2, MGC53152; prolyl-tRNA synthetase 2, mitochondrial
(putative); K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=466
Score = 33.9 bits (76), Expect = 0.96, Method: Compositional matrix adjust.
Identities = 36/161 (22%), Positives = 68/161 (42%), Gaps = 20/161 (12%)
Query 61 RGIQAATSHLLGQNFSRMFSIEFEDEKGAKQLVHQTSWGCTTRSL---GVMIMTHGDD-- 115
+GI+ + LG +S +F D + L +G L + ++++ DD
Sbjct 294 KGIEIGHTFYLGTKYSHVFKANCYDSQKTPFLAEMGCYGIGVSRLLAASLEVLSNEDDIH 353
Query 116 -KGLVLPPRVVAVQAVIIPIIFKETGGMEI----VAKCRELERLLNAAGVR--VKVDDRT 168
GL+ P +V +I + G EI VA+ + + + G+R +DDR+
Sbjct 354 WPGLIAPYQVC--------LISPKKGSKEIEAISVAETLYDDIIQSVCGLRDETVLDDRS 405
Query 169 NYTPGWKFNDWELKGVPLRLEIGPRDVESCQTRVVRRDTSE 209
T G + + + G P + IG + +E+ VR ++
Sbjct 406 YLTIGKRVKEAHMMGYPYVIVIGKKALETPSLFEVRSHNTD 446
> ath:AT1G35150 hypothetical protein
Length=459
Score = 33.5 bits (75), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 33/133 (24%), Positives = 47/133 (35%), Gaps = 25/133 (18%)
Query 154 LLNAAGVRVKVDDRTNYTPGWKFNDWELKGVPLRLEIGPRDVESCQTR------------ 201
LL G+ + D + Y +NDW L+ G D +C TR
Sbjct 80 LLVEKGLATRHDKKGGYLATSGYNDWRNLSKRLKEHKGSHDHITCMTRRAELESRLQKNK 139
Query 202 VVRRDTSEA---RNVPWAEAASTIPAMLETMQKDLF----------NKAKAKFEASIEQI 248
+ + EA N+ W E I A+++T K+ F + IE I
Sbjct 140 TIDKHAQEAINKDNIHWREVLLRIIALVKTHAKNNLAFRGKNEKVGQDRNGNFLSFIEMI 199
Query 249 TTFDEVMPALNRR 261
FD VM RR
Sbjct 200 AEFDVVMREHIRR 212
> dre:562478 zufsp, MGC158611, im:7145879, zgc:158611; zinc finger
with UFM1-specific peptidase domain
Length=608
Score = 32.7 bits (73), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 31/124 (25%), Positives = 49/124 (39%), Gaps = 26/124 (20%)
Query 110 MTHGDDKGLVLPPRVVAVQAVIIPIIFKETG------GMEIVAKCR-------------E 150
TH +G LPP+V VQ + P+ + G GME A + E
Sbjct 492 FTHSTSRGARLPPKV--VQTTLPPVYLQHQGHSRSIIGMEEKANGKLCLLFFDPGVTPAE 549
Query 151 LERLLNAAGVRVKVDDRTNYTPGWKFNDWELKGVPLRLEIGPRDVESCQTRVVRRDTSEA 210
++++L+ V V YT G K +++ V G E Q+R++ T A
Sbjct 550 MKKVLSQDTVTSMVQRMRKYTSGLKHKQYQVVIVE-----GVLTAEEKQSRILNSRTLRA 604
Query 211 RNVP 214
+P
Sbjct 605 EKIP 608
> dre:449661 tars, ThrRS, wu:fb07c05, wu:fb39c12, zgc:92586; threonyl-tRNA
synthetase (EC:6.1.1.3); K01868 threonyl-tRNA synthetase
[EC:6.1.1.3]
Length=624
Score = 32.7 bits (73), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 40/168 (23%), Positives = 64/168 (38%), Gaps = 16/168 (9%)
Query 43 FAGGKKTTTIEGIIPDTGRGIQAATSHLLGQ---NFSRMFSIEFEDEKGAKQLVHQTSWG 99
F G K I+ I GR Q AT L Q F+ F D+K ++H+ G
Sbjct 440 FYGPKIDIQIKDAI---GRYHQCATIQLDFQLPIRFNLTFVSHDGDDKKRPVIIHRAILG 496
Query 100 CTTRSLGVMIMTHGDDKGLVLPPRVVAVQAVIIPIIFKETGGMEIVAKCRELERLLNAAG 159
R + ++ +G L L PR Q +++PI G + +++ + G
Sbjct 497 SVERMIAILTENYGGKWPLWLSPR----QVMVVPI------GPTCDEYAQRVQKEFHKVG 546
Query 160 VRVKVDDRTNYTPGWKFNDWELKGVPLRLEIGPRDVESCQTRVVRRDT 207
+ VD T K + +L L +G ++ S V RD
Sbjct 547 LMTDVDLDPGCTLNKKIRNAQLAQYNFILVVGEKEKSSDTVNVRTRDN 594
> sce:YDR476C Putative protein of unknown function; green fluorescent
protein (GFP)-fusion protein localizes to the endoplasmic
reticulum; YDR476C is not an essential gene
Length=224
Score = 32.3 bits (72), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 37/146 (25%), Positives = 59/146 (40%), Gaps = 29/146 (19%)
Query 11 TLVLEILELYRQWYEDYLAV----PVIKGEK------------SENEKFAGGKKTTTIEG 54
+L++ I + ++ ED LAV P+ K K + KF + IE
Sbjct 4 SLIVSINDTHKLGLEDCLAVFGHVPITKAVKHVRLTEIDTQTSTFTLKFLHTETGQNIEK 63
Query 55 II----PDTGRGIQAATSHLLGQNFSRMFSIEFEDEKGAKQLVHQTSWGCTTRSLGVMIM 110
II DTG + AT + Q F++MF I E K + + + CT L +++
Sbjct 64 IIYFIDNDTGNDTRTATG--IKQIFNKMFRIAAEKRKLSLIQIDTVEYPCTLVDLLILV- 120
Query 111 THGDDKGLVLPPRVVAVQAVIIPIIF 136
G+ LPP + + I F
Sbjct 121 ------GVALPPLCYLYRPALHAIFF 140
> hsa:57731 SPTBN4, KIAA1642, QV, SPNB4, SPTBN3; spectrin, beta,
non-erythrocytic 4; K06115 spectrin beta
Length=2564
Score = 31.2 bits (69), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 18/53 (33%), Positives = 27/53 (50%), Gaps = 0/53 (0%)
Query 7 EEAYTLVLEILELYRQWYEDYLAVPVIKGEKSENEKFAGGKKTTTIEGIIPDT 59
E+ + V E+ L RQW D LAV + GE E + G K+ + +PD+
Sbjct 860 EQLFAEVTEVAALRRQWLRDALAVYRMFGEVHACELWIGEKEQWLLSMRVPDS 912
> tgo:TGME49_085840 hypothetical protein
Length=571
Score = 30.8 bits (68), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 18/39 (46%), Positives = 21/39 (53%), Gaps = 0/39 (0%)
Query 273 ETETQIKRETQRLSEIQARENADGMTGAMKPLCIPFDQP 311
E E IK TQRLS A N + TGA +P+ I D P
Sbjct 169 EKEQLIKELTQRLSGPLADSNGNVPTGAARPIAIEVDGP 207
> hsa:360200 TMPRSS9, FLJ16193; transmembrane protease, serine
9; K09640 transmembrane protease, serine 9 [EC:3.4.21.-]
Length=1059
Score = 30.8 bits (68), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 11/23 (47%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 301 MKPLCIPFDQPPMPEGTRCFFTG 323
++P+C+P P P+GTRC TG
Sbjct 931 VRPICLPEPAPRPPDGTRCVITG 953
> xla:446392 b9d1, MGC83744; B9 protein domain 1
Length=198
Score = 30.8 bits (68), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 24/88 (27%), Positives = 37/88 (42%), Gaps = 11/88 (12%)
Query 26 DYLAVPVIKGEKSENEKFAGGKKTTTIEGIIPDTGRGIQAATSHLLGQNFSRMFSIEFED 85
D V++G + + F G+ T TI +P++ +Q TS +G+ EF D
Sbjct 92 DVFGNDVVRGYGAVHLPFTPGRHTRTIPMFVPESSSRLQRFTSWFMGRR------PEFTD 145
Query 86 EKGAKQ-----LVHQTSWGCTTRSLGVM 108
K Q + S GC T S V+
Sbjct 146 PKVVAQGEGREVTRVRSQGCVTVSFNVV 173
Lambda K H
0.319 0.134 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 14156784820
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40