bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0524_orf2
Length=122
Score E
Sequences producing significant alignments: (Bits) Value
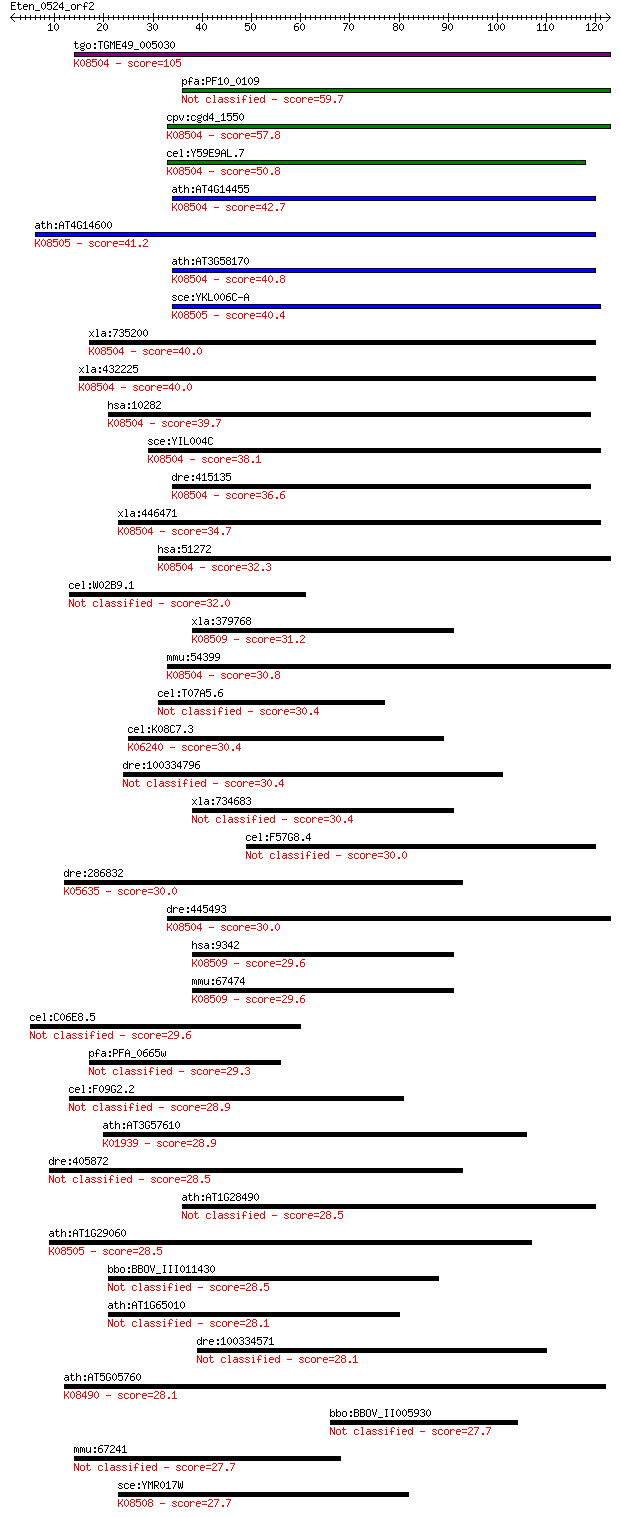
tgo:TGME49_005030 hypothetical protein ; K08504 blocked early ... 105 5e-23
pfa:PF10_0109 conserved Plasmodium protein 59.7 2e-09
cpv:cgd4_1550 hypothetical protein ; K08504 blocked early in t... 57.8 7e-09
cel:Y59E9AL.7 hypothetical protein; K08504 blocked early in tr... 50.8 1e-06
ath:AT4G14455 ATBET12; SNAP receptor/ protein transporter; K08... 42.7 3e-04
ath:AT4G14600 hypothetical protein; K08505 protein transport p... 41.2 7e-04
ath:AT3G58170 BS14A; BS14A (BET1P/SFT1P-LIKE PROTEIN 14A); SNA... 40.8 0.001
sce:YKL006C-A SFT1; Intra-Golgi V-SNARE, required for transpor... 40.4 0.001
xla:735200 hypothetical protein MGC131356; K08504 blocked earl... 40.0 0.002
xla:432225 bet1, MGC84493; blocked early in transport 1 homolo... 40.0 0.002
hsa:10282 BET1, DKFZp781C0425, HBET1; blocked early in transpo... 39.7 0.003
sce:YIL004C BET1, SLY12; Bet1p; K08504 blocked early in transp... 38.1 0.007
dre:415135 bet1, si:ch211-262k23.5, zgc:86622; blocked early i... 36.6 0.017
xla:446471 bet1l, MGC79029; blocked early in transport 1 homol... 34.7 0.077
hsa:51272 BET1L, BET1L1, GOLIM3, GS15, HSPC197; blocked early ... 32.3 0.40
cel:W02B9.1 hmr-1; HaMmeRhead embryonic lethal family member (... 32.0 0.46
xla:379768 snap29, MGC52674; synaptosomal-associated protein, ... 31.2 0.81
mmu:54399 Bet1l, 2610021K23Rik, Gs15; blocked early in transpo... 30.8 1.2
cel:T07A5.6 unc-69; UNCoordinated family member (unc-69) 30.4 1.4
cel:K08C7.3 epi-1; abnormal EPIthelia family member (epi-1); K... 30.4 1.4
dre:100334796 Pre-mRNA-processing factor 39-like 30.4 1.4
xla:734683 hypothetical protein MGC116490 30.4 1.4
cel:F57G8.4 srw-87; Serpentine Receptor, class W family member... 30.0 1.7
dre:286832 lamc1, sly; laminin, gamma 1; K05635 laminin, gamma 1 30.0
dre:445493 bet1l; zgc:100789; K08504 blocked early in transport 1 30.0
hsa:9342 SNAP29, CEDNIK, FLJ21051, SNAP-29; synaptosomal-assoc... 29.6 2.2
mmu:67474 Snap29, 1300018G05Rik, AI891940, AU020222, BB131856,... 29.6 2.3
cel:C06E8.5 hypothetical protein 29.6 2.4
pfa:PFA_0665w DBL containing protein, unknown function 29.3 3.2
cel:F09G2.2 hypothetical protein 28.9 4.2
ath:AT3G57610 ADSS; ADSS (ADENYLOSUCCINATE SYNTHASE); adenylos... 28.9 4.3
dre:405872 MGC85889; zgc:85889 28.5 4.7
ath:AT1G28490 SYP61; SYP61 (SYNTAXIN OF PLANTS 61); SNAP receptor 28.5 5.1
ath:AT1G29060 hypothetical protein; K08505 protein transport p... 28.5 5.4
bbo:BBOV_III011430 17.m07975; hypothetical protein 28.5 5.6
ath:AT1G65010 hypothetical protein 28.1 6.5
dre:100334571 neuronal pentraxin I-like 28.1 6.5
ath:AT5G05760 SYP31; SYP31 (SYNTAXIN OF PLANTS 31); SNAP recep... 28.1 7.6
bbo:BBOV_II005930 18.m06491; rhomboid 4 27.7
mmu:67241 Smc6, 2810489L22Rik, 3830418C19Rik, AA990493, AU0187... 27.7 8.6
sce:YMR017W SPO20, DBI9; Meiosis-specific subunit of the t-SNA... 27.7 8.8
> tgo:TGME49_005030 hypothetical protein ; K08504 blocked early
in transport 1
Length=107
Score = 105 bits (261), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 56/111 (50%), Positives = 79/111 (71%), Gaps = 9/111 (8%)
Query 14 MMRKKENGNEWKQI--KNLESLEAENDSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGT 71
MMRKKE G E ++ + ++++E END+ I DLE++ LSLAMRDEVQ SN+LL
Sbjct 1 MMRKKE-GYERRRFEQRGMDAMEEENDACIVDLESK------LSLAMRDEVQESNNLLEG 53
Query 72 MVGRLEGVRVSVRNSVRRMDRMFKSGGGWHMLHLAAFVLFIFLIFYFFFAK 122
M G L+GVR S+R+S++RM+ ++ GGWH +LA FV+ IF+IFYF + K
Sbjct 54 MAGGLDGVRNSIRSSIKRMENLWNQRGGWHTCYLALFVVVIFIIFYFLYGK 104
> pfa:PF10_0109 conserved Plasmodium protein
Length=114
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 34/87 (39%), Positives = 53/87 (60%), Gaps = 0/87 (0%)
Query 36 ENDSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGTMVGRLEGVRVSVRNSVRRMDRMFK 95
END+YI DLE +VQ LK + +RDEV+TSNSLL + R+E V + RR+ + +
Sbjct 26 ENDNYILDLEKKVQSLKLIGSNLRDEVRTSNSLLDNLSDRMENVNKKLSGVYRRVKNILR 85
Query 96 SGGGWHMLHLAAFVLFIFLIFYFFFAK 122
+ G +M++L F LF+ ++ + K
Sbjct 86 TKGNNYMMYLILFFLFLLFFMHYLYRK 112
> cpv:cgd4_1550 hypothetical protein ; K08504 blocked early in
transport 1
Length=93
Score = 57.8 bits (138), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 28/90 (31%), Positives = 52/90 (57%), Gaps = 0/90 (0%)
Query 33 LEAENDSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGTMVGRLEGVRVSVRNSVRRMDR 92
LE +ND Y+ DLEA+VQ LK +S MR +V+ SN LL ++ + V ++ ++ ++
Sbjct 3 LEEQNDQYMIDLEAKVQDLKYISNTMRQQVKESNDLLSSLSEEMGSFGVLLKGTINKIKT 62
Query 93 MFKSGGGWHMLHLAAFVLFIFLIFYFFFAK 122
+ + H+ ++ F+ +FL+ Y F +
Sbjct 63 IGGTSSWKHIWIMSFFIFLVFLLMYILFKR 92
> cel:Y59E9AL.7 hypothetical protein; K08504 blocked early in
transport 1
Length=107
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 25/85 (29%), Positives = 49/85 (57%), Gaps = 0/85 (0%)
Query 33 LEAENDSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGTMVGRLEGVRVSVRNSVRRMDR 92
LE ND + L ++V LK +++A+ D+V+ N LL M + + +++++RR+
Sbjct 18 LERHNDDLVGGLSSKVAALKRVTIAIGDDVREQNRLLNDMDNDFDSSKGLLQSTMRRLGL 77
Query 93 MFKSGGGWHMLHLAAFVLFIFLIFY 117
+ ++GG + +L F LF+F + Y
Sbjct 78 VSRAGGKNMLCYLILFALFVFFVVY 102
> ath:AT4G14455 ATBET12; SNAP receptor/ protein transporter; K08504
blocked early in transport 1
Length=130
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 22/86 (25%), Positives = 45/86 (52%), Gaps = 0/86 (0%)
Query 34 EAENDSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGTMVGRLEGVRVSVRNSVRRMDRM 93
E +ND +++L+ RV LK ++ + +EV+ N LL + +++ R + ++ R +
Sbjct 36 ERDNDEALENLQDRVSFLKRVTGDIHEEVENHNRLLDKVGNKMDSARGIMSGTINRFKLV 95
Query 94 FKSGGGWHMLHLAAFVLFIFLIFYFF 119
F+ L A+ + +FLI Y+
Sbjct 96 FEKKSNRKSCKLIAYFVLLFLIMYYL 121
> ath:AT4G14600 hypothetical protein; K08505 protein transport
protein SFT1
Length=137
Score = 41.2 bits (95), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 23/114 (20%), Positives = 56/114 (49%), Gaps = 1/114 (0%)
Query 6 SSVRTPQRMMRKKENGNEWKQIKNLESLEAENDSYIDDLEARVQGLKELSLAMRDEVQTS 65
+ R+ + + + G+E Q++ ++ + ++ D I L +V+ LK ++ + E +
Sbjct 19 APYRSREGLSTRNAAGSEEIQLR-IDPMHSDLDDEITGLHGQVRQLKNIAQEIGSEAKFQ 77
Query 66 NSLLGTMVGRLEGVRVSVRNSVRRMDRMFKSGGGWHMLHLAAFVLFIFLIFYFF 119
L + L + V+N++R+++ G H++H+ F L +F + Y +
Sbjct 78 RDFLDELQMTLIRAQAGVKNNIRKLNMSIIRSGNNHIMHVVLFALLVFFVLYIW 131
> ath:AT3G58170 BS14A; BS14A (BET1P/SFT1P-LIKE PROTEIN 14A); SNAP
receptor/ protein transporter; K08504 blocked early in transport
1
Length=122
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 26/86 (30%), Positives = 45/86 (52%), Gaps = 0/86 (0%)
Query 34 EAENDSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGTMVGRLEGVRVSVRNSVRRMDRM 93
E EN+ ++ L+ RV LK LS + +EV T N +L M ++ R + ++ R +
Sbjct 35 EHENERALEGLQDRVILLKRLSGDINEEVDTHNRMLDRMGNDMDSSRGFLSGTMDRFKTV 94
Query 94 FKSGGGWHMLHLAAFVLFIFLIFYFF 119
F++ ML L A + +FL+ Y+
Sbjct 95 FETKSSRRMLTLVASFVGLFLVIYYL 120
> sce:YKL006C-A SFT1; Intra-Golgi V-SNARE, required for transport
of proteins between an early and a later Golgi compartment;
K08505 protein transport protein SFT1
Length=97
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/88 (27%), Positives = 43/88 (48%), Gaps = 4/88 (4%)
Query 34 EAENDSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGTMVGRLEGVRVSVRNSVRRMDRM 93
E+ ND ++ L ++ + ++ + D + +S++ M L + ++NS R+ R
Sbjct 10 ESNNDRKLEGLANKLATFRNINQEIGDRAVSDSSVINQMTDSLGSMFTDIKNSSSRLTRS 69
Query 94 FKSGGG-WHMLHLAAFVLFIFLIFYFFF 120
K+G W M+ LA L IF I Y F
Sbjct 70 LKAGNSIWRMVGLA---LLIFFILYTLF 94
> xla:735200 hypothetical protein MGC131356; K08504 blocked early
in transport 1
Length=113
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/103 (22%), Positives = 48/103 (46%), Gaps = 0/103 (0%)
Query 17 KKENGNEWKQIKNLESLEAENDSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGTMVGRL 76
K G ++ E EN+ ++L+ + LK LS+ + +EV+ N +LG M
Sbjct 7 KHREGGPRTEMSGYSVYEEENERLTENLKMKASALKSLSIDIGNEVKYHNKMLGEMDSDF 66
Query 77 EGVRVSVRNSVRRMDRMFKSGGGWHMLHLAAFVLFIFLIFYFF 119
+ + ++ R+ + + + ++ F LF+F + Y+F
Sbjct 67 DSTGGLLGATMGRLRILSRGSQAKLLCYMMLFALFVFFVIYWF 109
> xla:432225 bet1, MGC84493; blocked early in transport 1 homolog;
K08504 blocked early in transport 1
Length=115
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/106 (21%), Positives = 52/106 (49%), Gaps = 1/106 (0%)
Query 15 MRKKENGNEWK-QIKNLESLEAENDSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGTMV 73
M+ +E G + ++ E EN+ ++L+ + LK LS+ + +EV+ N +LG M
Sbjct 6 MKHREGGPGPRTEMSGYSVYEEENERLTENLKMKTSALKSLSIDIGNEVKYHNKMLGEMD 65
Query 74 GRLEGVRVSVRNSVRRMDRMFKSGGGWHMLHLAAFVLFIFLIFYFF 119
+ + +++ R+ + + + ++ F F+F + Y+F
Sbjct 66 SDFDSTGGLLGSTMGRLKILSRGSQAKLLCYMMLFAFFVFFVIYWF 111
> hsa:10282 BET1, DKFZp781C0425, HBET1; blocked early in transport
1 homolog (S. cerevisiae); K08504 blocked early in transport
1
Length=118
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 23/98 (23%), Positives = 46/98 (46%), Gaps = 0/98 (0%)
Query 21 GNEWKQIKNLESLEAENDSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGTMVGRLEGVR 80
GN + E EN+ + L ++V +K LS+ + EV+T N LL M + +
Sbjct 16 GNYGYANSGYSACEEENERLTESLRSKVTAIKSLSIEIGHEVKTQNKLLAEMDSQFDSTT 75
Query 81 VSVRNSVRRMDRMFKSGGGWHMLHLAAFVLFIFLIFYF 118
+ ++ ++ + + + ++ F LF+F I Y+
Sbjct 76 GFLGKTMGKLKILSRGSQTKLLCYMMLFSLFVFFIIYW 113
> sce:YIL004C BET1, SLY12; Bet1p; K08504 blocked early in transport
1
Length=142
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 22/92 (23%), Positives = 45/92 (48%), Gaps = 4/92 (4%)
Query 29 NLESLEAENDSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGTMVGRLEGVRVSVRNSVR 88
L SLE++++ + + R++ LK LSL M DE++ SN + + V ++ +
Sbjct 50 TLASLESQSEEQMGAMGQRIKALKSLSLKMGDEIRGSNQTIDQLGDTFHNTSVKLKRTFG 109
Query 89 RMDRMFKSGGGWHMLHLAAFVLFIFLIFYFFF 120
M M + G + + +++ F++ FF
Sbjct 110 NMMEMARRSG----ISIKTWLIIFFMVGVLFF 137
> dre:415135 bet1, si:ch211-262k23.5, zgc:86622; blocked early
in transport 1 homolog (S. cerevisiae); K08504 blocked early
in transport 1
Length=113
Score = 36.6 bits (83), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 22/85 (25%), Positives = 42/85 (49%), Gaps = 0/85 (0%)
Query 34 EAENDSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGTMVGRLEGVRVSVRNSVRRMDRM 93
E EN+ + L +V LK LS+ + +EV+ N +LG M + + ++ R+ +
Sbjct 24 EEENEHLQEGLRDKVHALKHLSIDIGNEVKIHNKMLGEMDSDFDSTGGLLGATMGRLKLL 83
Query 94 FKSGGGWHMLHLAAFVLFIFLIFYF 118
+ ++ F LF+FL+ Y+
Sbjct 84 SRGSQTKVYCYMLLFALFVFLVLYW 108
> xla:446471 bet1l, MGC79029; blocked early in transport 1 homolog-like;
K08504 blocked early in transport 1
Length=111
Score = 34.7 bits (78), Expect = 0.077, Method: Compositional matrix adjust.
Identities = 29/102 (28%), Positives = 43/102 (42%), Gaps = 4/102 (3%)
Query 23 EWKQIKNL----ESLEAENDSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGTMVGRLEG 78
+W ++ N E L+AEN D+L ++V LK L+L + E N L M
Sbjct 3 DWGRVPNSGAVDEMLDAENKRLADNLSSKVTRLKSLALDIDQEADDHNKYLDGMDSDFLS 62
Query 79 VRVSVRNSVRRMDRMFKSGGGWHMLHLAAFVLFIFLIFYFFF 120
V + SV+R M SG L V + L F ++
Sbjct 63 VTGLLGGSVKRFTGMALSGRDNRKLLCYVSVGLVALFFLLYY 104
> hsa:51272 BET1L, BET1L1, GOLIM3, GS15, HSPC197; blocked early
in transport 1 homolog (S. cerevisiae)-like; K08504 blocked
early in transport 1
Length=111
Score = 32.3 bits (72), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 25/94 (26%), Positives = 42/94 (44%), Gaps = 2/94 (2%)
Query 31 ESLEAENDSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGTMVGRLEGVRVSVRNSVRRM 90
E L+ EN D L ++V LK L+L + + + N L M + + SV+R
Sbjct 15 EILDRENKRMADSLASKVTRLKSLALDIDRDAEDQNRYLDGMDSDFTSMTSLLTGSVKRF 74
Query 91 DRMFKSGGGWHML--HLAAFVLFIFLIFYFFFAK 122
M +SG L +A ++ F I +F ++
Sbjct 75 STMARSGQDNRKLLCGMAVGLIVAFFILSYFLSR 108
> cel:W02B9.1 hmr-1; HaMmeRhead embryonic lethal family member
(hmr-1)
Length=2920
Score = 32.0 bits (71), Expect = 0.46, Method: Composition-based stats.
Identities = 18/54 (33%), Positives = 28/54 (51%), Gaps = 6/54 (11%)
Query 13 RMMRKKENGNEWKQIKNLESLEAEN-DSYI-----DDLEARVQGLKELSLAMRD 60
R K ENGNEW ++ +E L+ E ++Y D+ +RV K + +RD
Sbjct 1150 RQFNKFENGNEWVEVVIMEGLDYEQVNNYTLTLTATDMTSRVASTKTFVVEVRD 1203
> xla:379768 snap29, MGC52674; synaptosomal-associated protein,
29kDa; K08509 synaptosomal-associated protein, 29kDa
Length=257
Score = 31.2 bits (69), Expect = 0.81, Method: Compositional matrix adjust.
Identities = 12/53 (22%), Positives = 34/53 (64%), Gaps = 0/53 (0%)
Query 38 DSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGTMVGRLEGVRVSVRNSVRRM 90
D+ +DD+ + LK L+L ++ E+ + +LG + G+++ + ++++ + +R+
Sbjct 201 DNNLDDMSLGLGRLKNLALGLQTEIDEQDDILGRLTGKVDKMDLNIKATDKRI 253
> mmu:54399 Bet1l, 2610021K23Rik, Gs15; blocked early in transport
1 homolog (S. cerevisiae)-like; K08504 blocked early in
transport 1
Length=111
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 26/92 (28%), Positives = 41/92 (44%), Gaps = 2/92 (2%)
Query 33 LEAENDSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGTMVGRLEGVRVSVRNSVRRMDR 92
L+ EN D L ++V LK L+L + + + N L M V + SV+R
Sbjct 17 LDRENKRMADSLASKVTRLKSLALDIDRDTEDQNRYLDGMDSDFTSVTGLLTGSVKRFST 76
Query 93 MFKSG-GGWHMLHLAAFVLFI-FLIFYFFFAK 122
M +SG +L A VL + F I + ++
Sbjct 77 MARSGRDNRKLLCGMAVVLIVAFFILSYLLSR 108
> cel:T07A5.6 unc-69; UNCoordinated family member (unc-69)
Length=176
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 18/46 (39%), Positives = 26/46 (56%), Gaps = 6/46 (13%)
Query 31 ESLEAENDSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGTMVGRL 76
+ LE +N +DDL RV+ +KE SL +R E N +LG + L
Sbjct 52 QVLELQNT--LDDLSQRVESVKEESLKLRSE----NQVLGQYIQNL 91
> cel:K08C7.3 epi-1; abnormal EPIthelia family member (epi-1);
K06240 laminin, alpha 3/5
Length=3704
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 17/64 (26%), Positives = 38/64 (59%), Gaps = 10/64 (15%)
Query 25 KQIKNLESLEAENDSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGTMVGRLEGVRVSVR 84
++++ +E L+AE D+ I++ A++ E++ + + +NS +EG+R++ R
Sbjct 2587 EKVQEVEKLKAEIDANIEETRAKIS---EIAGKAEEITEKANS-------AMEGIRLARR 2636
Query 85 NSVR 88
NSV+
Sbjct 2637 NSVQ 2640
> dre:100334796 Pre-mRNA-processing factor 39-like
Length=707
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 23/80 (28%), Positives = 38/80 (47%), Gaps = 13/80 (16%)
Query 24 WKQIKNLESLEAENDSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGTMVGRLEGVRVSV 83
WK+ +LE N+ + E QGLK + L++ + N LLGT+ +++
Sbjct 119 WKKFADLERRAGHNEKAEEVCE---QGLKSIPLSVDLWIHYINLLLGTL-------NMNL 168
Query 84 RNSVRRMDRMFK---SGGGW 100
S RR+ +F+ S GW
Sbjct 169 PESTRRIRSVFEEAVSAAGW 188
> xla:734683 hypothetical protein MGC116490
Length=163
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 11/53 (20%), Positives = 33/53 (62%), Gaps = 0/53 (0%)
Query 38 DSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGTMVGRLEGVRVSVRNSVRRM 90
D+ +DD+ + LK L+L ++ E+ + +LG + +++ + ++++ + +R+
Sbjct 107 DNNLDDMSLGLSRLKNLALGLQTEIDDQDDMLGRLTNKVDKMDLNIKTTDKRV 159
> cel:F57G8.4 srw-87; Serpentine Receptor, class W family member
(srw-87)
Length=361
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 40/78 (51%), Gaps = 10/78 (12%)
Query 49 QGLKELSL-----AMRDEVQTSNSLLGTMVGRLEGVR-VSVRNSVRR-MDRMFKSGGGWH 101
Q L E SL +RD+ + ++ LG M L GVR V+VRN + + + + + GW
Sbjct 87 QSLLETSLDWFLYTLRDDFRRCSTWLGLM---LAGVRTVAVRNVMNQNYNHISQPAFGWK 143
Query 102 MLHLAAFVLFIFLIFYFF 119
+ + V F IFY+F
Sbjct 144 INFIVLGVSTFFSIFYYF 161
> dre:286832 lamc1, sly; laminin, gamma 1; K05635 laminin, gamma
1
Length=1593
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 20/81 (24%), Positives = 33/81 (40%), Gaps = 0/81 (0%)
Query 12 QRMMRKKENGNEWKQIKNLESLEAENDSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGT 71
Q ++ +N K E E + I DL Q KE+ + D + N L
Sbjct 1037 QNLIDSLDNTETTVSDKAFEDRLKEAEKTIMDLLEEAQASKEVDKGLLDRLNNINKTLNN 1096
Query 72 MVGRLEGVRVSVRNSVRRMDR 92
RL+ ++ +V N+ + DR
Sbjct 1097 QWNRLQNIKNTVDNTGAQADR 1117
> dre:445493 bet1l; zgc:100789; K08504 blocked early in transport
1
Length=110
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 20/92 (21%), Positives = 43/92 (46%), Gaps = 2/92 (2%)
Query 33 LEAENDSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGTMVGRLEGVRVSVRNSVRRMDR 92
L+AEN ++L ++V LK L+ + + + N+ L M + SV+R
Sbjct 16 LDAENKRMAENLASKVSRLKSLAYDIDKDAEEQNAYLDGMDSNFLSATGLLTGSVKRFST 75
Query 93 MFKSGGGWH--MLHLAAFVLFIFLIFYFFFAK 122
M +SG + +++ ++ F + Y+ ++
Sbjct 76 MVRSGRDNRKILCYVSVGLVVAFFLLYYLVSR 107
> hsa:9342 SNAP29, CEDNIK, FLJ21051, SNAP-29; synaptosomal-associated
protein, 29kDa; K08509 synaptosomal-associated protein,
29kDa
Length=258
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 12/53 (22%), Positives = 35/53 (66%), Gaps = 0/53 (0%)
Query 38 DSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGTMVGRLEGVRVSVRNSVRRM 90
DS +D+L + LK+++L M+ E++ + +L + +++ + V+++++ R++
Sbjct 203 DSNLDELSMGLGRLKDIALGMQTEIEEQDDILDRLTTKVDKLDVNIKSTERKV 255
> mmu:67474 Snap29, 1300018G05Rik, AI891940, AU020222, BB131856,
Gs32; synaptosomal-associated protein 29; K08509 synaptosomal-associated
protein, 29kDa
Length=260
Score = 29.6 bits (65), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 11/53 (20%), Positives = 35/53 (66%), Gaps = 0/53 (0%)
Query 38 DSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGTMVGRLEGVRVSVRNSVRRM 90
DS +D+L + LK+++L M+ E++ + +L + +++ + V+++++ +++
Sbjct 205 DSNLDELSVGLGHLKDIALGMQTEIEEQDDILDRLTTKVDKLDVNIKSTEKKV 257
> cel:C06E8.5 hypothetical protein
Length=579
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 13/55 (23%), Positives = 31/55 (56%), Gaps = 0/55 (0%)
Query 5 FSSVRTPQRMMRKKENGNEWKQIKNLESLEAENDSYIDDLEARVQGLKELSLAMR 59
F+++R P +M+ +E G ++ L L ++ + I ++E R++ L ++ L +
Sbjct 431 FTTLRAPAVIMQPEEKGGIHFSLRGLIRLVSDRNEPIGEMEIRIEALMKMKLTSK 485
> pfa:PFA_0665w DBL containing protein, unknown function
Length=2900
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 23/39 (58%), Gaps = 0/39 (0%)
Query 17 KKENGNEWKQIKNLESLEAENDSYIDDLEARVQGLKELS 55
K EN + K+ KN+ S+ N+ Y D++E R KE++
Sbjct 2715 KNENLHTEKKKKNINSIRDNNNDYYDNVENRFLKRKEIA 2753
> cel:F09G2.2 hypothetical protein
Length=319
Score = 28.9 bits (63), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 22/74 (29%), Positives = 39/74 (52%), Gaps = 10/74 (13%)
Query 13 RMMRKKENGNEWKQIKNLE-SLEAEND---SYIDDLEARVQGLKELSLAMRDEVQTSNSL 68
+M + N EW +KNL+ ++ +ND +Y+D++E V G S ++ T N L
Sbjct 126 KMTPEDLNAKEWNLVKNLDWNVAVKNDEFENYLDNMEKWVAG----SFVEKNGFMTYNEL 181
Query 69 --LGTMVGRLEGVR 80
L +MV ++ V+
Sbjct 182 LQLSSMVPIMDIVK 195
> ath:AT3G57610 ADSS; ADSS (ADENYLOSUCCINATE SYNTHASE); adenylosuccinate
synthase (EC:6.3.4.4); K01939 adenylosuccinate synthase
[EC:6.3.4.4]
Length=490
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 18/86 (20%), Positives = 38/86 (44%), Gaps = 0/86 (0%)
Query 20 NGNEWKQIKNLESLEAENDSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGTMVGRLEGV 79
NG ++++++L + D + D AR QG K +R+EV+ + +
Sbjct 210 NGIRVGDLRHMDTLPQKLDLLLSDAAARFQGFKYTPEMLREEVEAYKRYADRLEPYITDT 269
Query 80 RVSVRNSVRRMDRMFKSGGGWHMLHL 105
+ +S+ + ++ GG ML +
Sbjct 270 VHFINDSISQKKKVLVEGGQATMLDI 295
> dre:405872 MGC85889; zgc:85889
Length=432
Score = 28.5 bits (62), Expect = 4.7, Method: Composition-based stats.
Identities = 25/91 (27%), Positives = 43/91 (47%), Gaps = 15/91 (16%)
Query 9 RTPQRMMRKKENGNEWKQIKN---LESLEAENDSYIDDLE----ARVQGLKELSLAMRDE 61
+T Q + ++ EN ++ + N SL+ S IDDLE +RV GL+E MR+E
Sbjct 135 QTLQSLKQRLENLEQFSRNNNSVQANSLKDLLQSKIDDLEKQVLSRVNGLEEGKPGMRNE 194
Query 62 VQTSNSLLGTMVGRLEGVRVSVRNSVRRMDR 92
+ GR+E S+ + +++
Sbjct 195 SEQR--------GRVESTLTSLHQRITDLEK 217
> ath:AT1G28490 SYP61; SYP61 (SYNTAXIN OF PLANTS 61); SNAP receptor
Length=206
Score = 28.5 bits (62), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 14/84 (16%), Positives = 38/84 (45%), Gaps = 0/84 (0%)
Query 36 ENDSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGTMVGRLEGVRVSVRNSVRRMDRMFK 95
+ D +D+L VQ + + L + DE+ ++ + ++ + + +++ + K
Sbjct 119 QQDEELDELSKSVQRIGGVGLTIHDELVAQERIIDELDTEMDSTKNRLEFVQKKVGMVMK 178
Query 96 SGGGWHMLHLAAFVLFIFLIFYFF 119
G + + F+L +F+I +
Sbjct 179 KAGAKGQMMMICFLLVLFIILFVL 202
> ath:AT1G29060 hypothetical protein; K08505 protein transport
protein SFT1
Length=134
Score = 28.5 bits (62), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 19/98 (19%), Positives = 50/98 (51%), Gaps = 1/98 (1%)
Query 9 RTPQRMMRKKENGNEWKQIKNLESLEAENDSYIDDLEARVQGLKELSLAMRDEVQTSNSL 68
R+ + + + +G+E Q++ ++ + ++ D I L +V+ LK ++ + E ++
Sbjct 19 RSREGLSTRNASGSEEIQLR-IDPMHSDLDDEILGLHGQVRQLKNIAQEIGSEAKSQRDF 77
Query 69 LGTMVGRLEGVRVSVRNSVRRMDRMFKSGGGWHMLHLA 106
L + L + V+N++R+++ G H++H+
Sbjct 78 LDELQMTLIRAQAGVKNNIRKLNLSIIRSGNNHIMHVV 115
> bbo:BBOV_III011430 17.m07975; hypothetical protein
Length=666
Score = 28.5 bits (62), Expect = 5.6, Method: Composition-based stats.
Identities = 16/67 (23%), Positives = 33/67 (49%), Gaps = 9/67 (13%)
Query 21 GNEWKQIKNLESLEAENDSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGTMVGRLEGVR 80
GN K ++ ++S+ +E +Y++DLE L+ + V TS+ + L +
Sbjct 506 GNNVKSVEKMDSVHSEGGNYVNDLEC---------LSFDNPVTTSSQDINVASAALSADK 556
Query 81 VSVRNSV 87
++R S+
Sbjct 557 YNIRTSL 563
> ath:AT1G65010 hypothetical protein
Length=1345
Score = 28.1 bits (61), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 20/69 (28%), Positives = 34/69 (49%), Gaps = 11/69 (15%)
Query 21 GNEWKQIKNLESLEAENDSYIDDLEARVQGLKELSLAMR----------DEVQTSNSLLG 70
G E +K +E L EN++ +D++ A +Q + E S +R DE+ T+N L
Sbjct 789 GREASHLKKIEELSKENENLVDNV-ANMQNIAEESKDLREREVAYLKKIDELSTANGTLA 847
Query 71 TMVGRLEGV 79
V L+ +
Sbjct 848 DNVTNLQNI 856
> dre:100334571 neuronal pentraxin I-like
Length=253
Score = 28.1 bits (61), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 20/75 (26%), Positives = 35/75 (46%), Gaps = 12/75 (16%)
Query 39 SYIDDLE----ARVQGLKELSLAMRDEVQTSNSLLGTMVGRLEGVRVSVRNSVRRMDRMF 94
S IDDLE +RV GL+E MR+E + GR+E S+ + +++ F
Sbjct 168 SKIDDLEKQVLSRVNGLEEGKPGMRNESE--------QRGRVESTLTSLHQRITDLEKAF 219
Query 95 KSGGGWHMLHLAAFV 109
+ + + L + +
Sbjct 220 RQQSSPYTVALDSCL 234
> ath:AT5G05760 SYP31; SYP31 (SYNTAXIN OF PLANTS 31); SNAP receptor;
K08490 syntaxin 5
Length=336
Score = 28.1 bits (61), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 25/112 (22%), Positives = 50/112 (44%), Gaps = 14/112 (12%)
Query 12 QRMMRKKENGNEWKQIK--NLESLEAENDSYIDDLEARVQGLKELSLAMRDEVQTSNSLL 69
Q+ + K+EN ++ + + ++ES E L V EL++ + D + S
Sbjct 237 QQTVPKQENYSQSRAVALHSVESRITELSGIFPQLATMVTQQGELAIRIDDNMDES---- 292
Query 70 GTMVGRLEGVRVSVRNSVRRMDRMFKSGGGWHMLHLAAFVLFIFLIFYFFFA 121
+ +EG R ++ + R+ S W M+ + A ++ ++F FF A
Sbjct 293 ---LVNVEGARSALLQHLTRI-----SSNRWLMMKIFAVIILFLIVFLFFVA 336
> bbo:BBOV_II005930 18.m06491; rhomboid 4
Length=783
Score = 27.7 bits (60), Expect = 8.3, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 21/39 (53%), Gaps = 1/39 (2%)
Query 66 NSLLGTMVGRLEGVRVSVR-NSVRRMDRMFKSGGGWHML 103
N +GT+VG L + + S R + +F GG WH+L
Sbjct 464 NYRIGTIVGALSANTIRIYGESSRLLTSIFLHGGRWHLL 502
> mmu:67241 Smc6, 2810489L22Rik, 3830418C19Rik, AA990493, AU018782,
AW742439, KIAA4103, MGC96146, SMC-6, Smc6l1, mKIAA4103;
structural maintenance of chromosomes 6
Length=1097
Score = 27.7 bits (60), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 16/54 (29%), Positives = 34/54 (62%), Gaps = 6/54 (11%)
Query 14 MMRKKENGNEWKQIKNLESLEAENDSYIDDLEARVQGLKELSLAMRDEVQTSNS 67
M ++KEN ++NL+SL+ E ++ D ++ ++ L EL+ ++DE+ ++S
Sbjct 765 MEQQKEN------MENLKSLKIEAENKYDTIKLKINQLSELADPLKDELNLADS 812
> sce:YMR017W SPO20, DBI9; Meiosis-specific subunit of the t-SNARE
complex, required for prospore membrane formation during
sporulation; similar to but not functionally redundant with
Sec9p; SNAP-25 homolog; K08508 synaptosomal-associated protein,
23kDa
Length=397
Score = 27.7 bits (60), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 17/62 (27%), Positives = 32/62 (51%), Gaps = 8/62 (12%)
Query 23 EWKQIK---NLESLEAENDSYIDDLEARVQGLKELSLAMRDEVQTSNSLLGTMVGRLEGV 79
+W++++ NLE+++ E D +D L + ++ K S+ TS +L + R G
Sbjct 150 QWRKVESQYNLENVQPEEDEIVDRLRSEIRSTKLKSVKT-----TSRTLEKAIEARCTGK 204
Query 80 RV 81
RV
Sbjct 205 RV 206
Lambda K H
0.323 0.136 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2008132680
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40