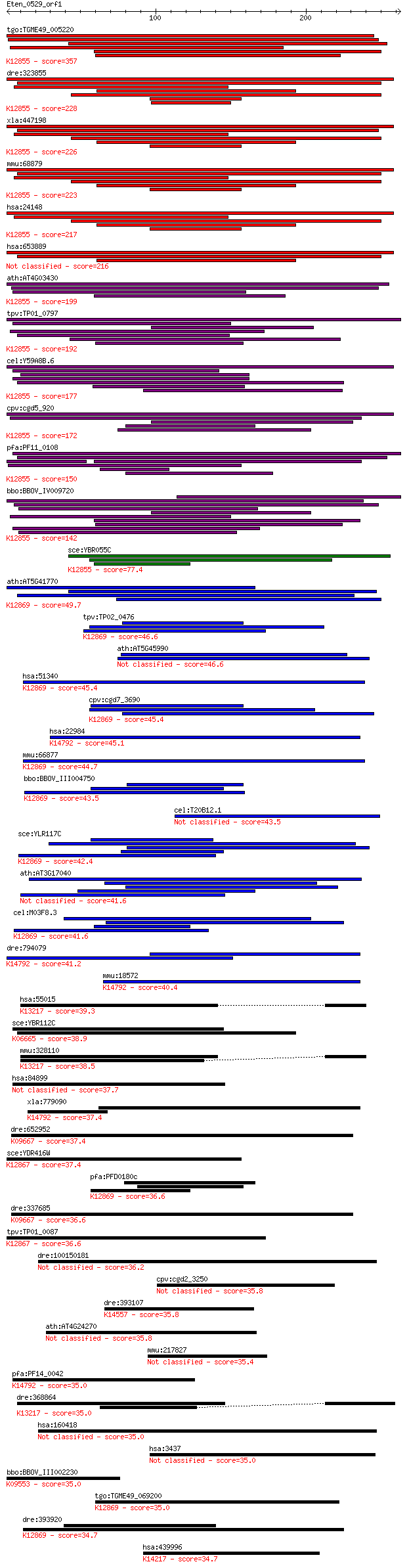
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0529_orf1
Length=262
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_005220 U5 snRNP-associated 102 kDa protein, putativ... 357 2e-98
dre:323855 c20orf14, fc12b02, prpf6, wu:fa05f07, wu:fc12b02, z... 228 2e-59
xla:447198 prpf6, MGC80263; PRP6 pre-mRNA processing factor 6 ... 226 6e-59
mmu:68879 Prpf6, 1190003A07Rik, 2610031L17Rik, ANT-1, MGC11655... 223 5e-58
hsa:24148 PRPF6, ANT-1, C20orf14, Prp6, TOM, U5-102K, hPrp6; P... 217 4e-56
hsa:653889 pre-mRNA-processing factor 6-like 216 6e-56
ath:AT4G03430 EMB2770 (EMBRYO DEFECTIVE 2770); RNA splicing fa... 199 7e-51
tpv:TP01_0797 hypothetical protein; K12855 pre-mRNA-processing... 192 1e-48
cel:Y59A8B.6 hypothetical protein; K12855 pre-mRNA-processing ... 177 3e-44
cpv:cgd5_920 Pre-mRNA splicing factor Pro1/Prp6. HAT repeat pr... 172 8e-43
pfa:PF11_0108 U5 snRNP-associated protein, putative; K12855 pr... 150 6e-36
bbo:BBOV_IV009720 23.m06014; u5 snRNP-associated subunit, puta... 142 2e-33
sce:YBR055C PRP6, RNA6, TSM7269; Prp6p; K12855 pre-mRNA-proces... 77.4 5e-14
ath:AT5G41770 crooked neck protein, putative / cell cycle prot... 49.7 1e-05
tpv:TP02_0476 crooked neck protein; K12869 crooked neck 46.6 8e-05
ath:AT5G45990 crooked neck protein, putative / cell cycle prot... 46.6 1e-04
hsa:51340 CRNKL1, CLF, CRN, Clf1, HCRN, MSTP021, SYF3; crooked... 45.4 2e-04
cpv:cgd7_3690 crooked neck protein HAT repeats ; K12869 crooke... 45.4 2e-04
hsa:22984 PDCD11, ALG-4, ALG4, KIAA0185, NFBP, RRP5; programme... 45.1 2e-04
mmu:66877 Crnkl1, 1200013P10Rik, 5730590A01Rik, C80326, crn; C... 44.7 3e-04
bbo:BBOV_III004750 17.m07426; tetratricopeptide repeat domain ... 43.5 7e-04
cel:T20B12.1 hypothetical protein 43.5 8e-04
sce:YLR117C CLF1, NTC77, SYF3; Clf1p; K12869 crooked neck 42.4 0.002
ath:AT3G17040 HCF107; HCF107 (HIGH CHLOROPHYLL FLUORESCENT 107... 41.6 0.003
cel:M03F8.3 hypothetical protein; K12869 crooked neck 41.6 0.003
dre:794079 pdcd11, MGC162501, cb680, im:7148359, sb:cb680, wu:... 41.2 0.004
mmu:18572 Pdcd11, 1110021I22Rik, ALG-4, Pdcd7, mKIAA0185; prog... 40.4 0.007
hsa:55015 PRPF39, FLJ11128, FLJ20666, FLJ45460, MGC149842, MGC... 39.3 0.016
sce:YBR112C CYC8, CRT8, SSN6; Cyc8p; K06665 glucose repression... 38.9 0.020
mmu:328110 Prpf39, FLJ11128, MGC37077, Srcs1; PRP39 pre-mRNA p... 38.5 0.022
hsa:84899 TMTC4, FLJ14624, FLJ22153; transmembrane and tetratr... 37.7 0.039
xla:779090 pdcd11, alg-4, alg4, nfbp, rrp5; programmed cell de... 37.4 0.050
dre:652952 ogt.2, im:7146393, ogtl, wu:fp46c04, wu:fr75f09; O-... 37.4 0.051
sce:YDR416W SYF1, NTC90; Member of the NineTeen Complex (NTC) ... 37.4 0.063
pfa:PFD0180c CGI-201 protein, short form; K12869 crooked neck 36.6 0.10
dre:337685 ogt.1, fm81g08, ogt, wu:fc12b01, wu:fm81g08; O-link... 36.6 0.10
tpv:TP01_0087 adapter protein; K12867 pre-mRNA-splicing factor... 36.6 0.10
dre:100150181 tmtc3; transmembrane and tetratricopeptide repea... 36.2 0.13
cpv:cgd2_3250 3x TPR domain-containing protein 35.8 0.15
dre:393107 utp6, MGC174851, MGC55648, zgc:55648; UTP6, small s... 35.8 0.17
ath:AT4G24270 RNA recognition motif (RRM)-containing protein 35.8 0.18
mmu:217827 6720454P05Rik; cDNA sequence BC002230 35.4 0.22
pfa:PF14_0042 U3 small nucleolar ribonucleoprotein, U3 snoRNP,... 35.0 0.25
dre:368864 prpf39, fa10d07, si:dz261o22.3, wu:fa10d07; PRP39 p... 35.0 0.26
hsa:160418 TMTC3, DKFZp686C0968, DKFZp686M1969, DKFZp686O22167... 35.0 0.28
hsa:3437 IFIT3, CIG-49, GARG-49, IFI60, IFIT4, IRG2, ISG60, RI... 35.0 0.31
bbo:BBOV_III002230 17.m07215; tetratricopeptide repeat domain ... 35.0 0.31
tgo:TGME49_069200 crooked neck-like protein 1, putative ; K128... 35.0 0.31
dre:393920 crnkl1, MGC55327, zgc:55327; crooked neck pre-mRNA ... 34.7 0.32
hsa:439996 IFIT1B, DKFZp781M1841, IFIT1L, bA149I23.6; interfer... 34.7 0.35
> tgo:TGME49_005220 U5 snRNP-associated 102 kDa protein, putative
; K12855 pre-mRNA-processing factor 6
Length=985
Score = 357 bits (917), Expect = 2e-98, Method: Compositional matrix adjust.
Identities = 164/245 (66%), Positives = 210/245 (85%), Gaps = 1/245 (0%)
Query 1 RQNINTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDL 60
R ++NT VW+QS+QLERQ+GDYDAAIAL +EA+K H KLWM+ GQLH EHPT++D
Sbjct 735 RAHVNTQKVWIQSVQLERQVGDYDAAIALCEEALKSHAECPKLWMIGGQLHREHPTKKDE 794
Query 61 ARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQ 120
+A +VF+ GTV C RSVPLWLC V+C+R Q W+ ARA+LEKAKLR P +P+LW A ++
Sbjct 795 EKAAEVFQRGTVVCCRSVPLWLCAVDCQREQGKWSVARAILEKAKLRNPKNPDLWHAAIR 854
Query 121 IEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCENDVNVIH 180
IE+E+GN+++AQHVA+KA+Q CPN+G++WAEAIFLEE++AQTH+AVDALTKCENDV+++
Sbjct 855 IEVEAGNKQMAQHVASKAVQECPNSGLVWAEAIFLEEKSAQTHKAVDALTKCENDVHLVL 914
Query 181 AVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELEN-GGEKECIDIINRCALA 239
AVA LFW++ KI+KARKW+NRSVTLD S GDAWA++LAFELEN GGEKEC +IIN+ +LA
Sbjct 915 AVACLFWKEGKISKARKWLNRSVTLDASFGDAWAAFLAFELENGGGEKECRNIINKASLA 974
Query 240 QPNRG 244
QPNRG
Sbjct 975 QPNRG 979
Score = 55.1 bits (131), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 58/287 (20%), Positives = 112/287 (39%), Gaps = 45/287 (15%)
Query 2 QNINTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLA 61
+ +N +W + + G A AL A+++ +T LW+ L +H T +DL
Sbjct 601 EGMNAKRIWKEDAEEALSRGSVATARALYTCAIERLKTKKSLWLALADLETKHGTTQDLE 660
Query 62 RAIDVFREGTVCCPRSVPLWLCW----------------------------------VNC 87
+ + + VCCP++ LWL V
Sbjct 661 K---LLAKAVVCCPQAEVLWLMLAKQHWLQGDVQAARKVLAEAFVHNENNEAISLAAVKL 717
Query 88 ERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQA---CPN 144
ER + RAR +L++ + V N+ ++W+ VQ+E + G+ + A + +A+++ CP
Sbjct 718 ERENHEFARARKILKRTRAHV-NTQKVWIQSVQLERQVGDYDAAIALCEEALKSHAECPK 776
Query 145 AGIIWAEAIFLEEENAQTHRAVDALTK----CENDVNVIHAVARLFWRDKKIAKARKWMN 200
+I + +A + + C V + K + AR +
Sbjct 777 LWMIGGQLHREHPTKKDEEKAAEVFQRGTVVCCRSVPLWLCAVDCQREQGKWSVARAILE 836
Query 201 RSVTLDPSLGDAWASYLAFELENGGEKECIDIINRCALAQPNRGLAW 247
++ +P D W + + E+E G ++ + ++ PN GL W
Sbjct 837 KAKLRNPKNPDLWHAAIRIEVEAGNKQMAQHVASKAVQECPNSGLVW 883
Score = 49.3 bits (116), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 50/224 (22%), Positives = 92/224 (41%), Gaps = 23/224 (10%)
Query 42 KLWMVRGQLHEEHPTQRDLARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVL 101
+LW L EE + L RA++ C P+SV +WL ++ A+ VL
Sbjct 471 RLWKEAVSLEEEKNARIMLTRAVE-------CVPQSVEIWLALARLS----SYEEAQKVL 519
Query 102 EKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFL---EEE 158
+A+ + P SPE+W+A ++E GN ++ + A+A G+ ++L EE
Sbjct 520 NEARKKCPTSPEIWVAACKLEETQGNLKMVDTIIARARDNLIARGVAQTRDVWLRLAEEA 579
Query 159 NAQTHRAV-DALTKCENDVNVIHAVARLFWRDK--------KIAKARKWMNRSVTLDPSL 209
A A A+ + V V A+ W++ +A AR ++ +
Sbjct 580 EASGFMATCQAIVRATMKVGVEGMNAKRIWKEDAEEALSRGSVATARALYTCAIERLKTK 639
Query 210 GDAWASYLAFELENGGEKECIDIINRCALAQPNRGLAWNKTTKQ 253
W + E ++G ++ ++ + + P + W KQ
Sbjct 640 KSLWLALADLETKHGTTQDLEKLLAKAVVCCPQAEVLWLMLAKQ 683
Score = 45.4 bits (106), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 56/206 (27%), Positives = 84/206 (40%), Gaps = 33/206 (16%)
Query 3 NINTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLAR 62
N + A W+ + +LE G AA L +Q +W+ +L + + LA+
Sbjct 371 NPHHAPGWIAAARLEELAGKLQAARELIATGCQQCPKSEDVWLEAARLEKPANAKAVLAK 430
Query 63 AID------------------------VFREGTVCCPRSVPLWLCWVNCERSQKNWNRAR 98
A+ V R+ P SV LW V+ E +KN AR
Sbjct 431 AVSVLPHSVRLWFDAYAREKDLDQRKRVLRKALEFIPNSVRLWKEAVSLE-EEKN---AR 486
Query 99 AVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEE 158
+L +A VP S E+WLAL ++ + E AQ V +A + CP + IW A LEE
Sbjct 487 IMLTRAVECVPQSVEIWLALARL----SSYEEAQKVLNEARKKCPTSPEIWVAACKLEET 542
Query 159 NAQTHRAVDALTKCENDVNVIHAVAR 184
+ VD + D + VA+
Sbjct 543 QGNL-KMVDTIIARARDNLIARGVAQ 567
Score = 38.5 bits (88), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 47/218 (21%), Positives = 79/218 (36%), Gaps = 31/218 (14%)
Query 59 DLARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLAL 118
D+ +A + + T P P W+ E AR ++ + P S ++WL
Sbjct 356 DIKKARTLLKSVTATNPHHAPGWIAAARLEELAGKLQAARELIATGCQQCPKSEDVWLEA 415
Query 119 VQIEIESGNREVA---------------------------QHVAAKAIQACPNAGIIWAE 151
++E + + V + V KA++ PN+ +W E
Sbjct 416 ARLEKPANAKAVLAKAVSVLPHSVRLWFDAYAREKDLDQRKRVLRKALEFIPNSVRLWKE 475
Query 152 AIFLEEENAQTHRAVDALTKCENDVNVIHAVARLFWRDKKIAKARKWMNRSVTLDPSLGD 211
A+ LEEE A+ V + A+ARL +A+K +N + P+ +
Sbjct 476 AVSLEEEKNARIMLTRAVECVPQSVEIWLALARL----SSYEEAQKVLNEARKKCPTSPE 531
Query 212 AWASYLAFELENGGEKECIDIINRCALAQPNRGLAWNK 249
W + E G K II R RG+A +
Sbjct 532 IWVAACKLEETQGNLKMVDTIIARARDNLIARGVAQTR 569
Score = 38.1 bits (87), Expect = 0.033, Method: Compositional matrix adjust.
Identities = 37/172 (21%), Positives = 75/172 (43%), Gaps = 19/172 (11%)
Query 60 LARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWN-----RARAVLEKAKLRVPNSPEL 114
L + +D TV P+ +L +N + Q + + +AR +L+ P+
Sbjct 321 LDKVMDNLSGQTVIDPKG---YLTDLNSMQLQSDADVADIKKARTLLKSVTATNPHHAPG 377
Query 115 WLALVQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCEN 174
W+A ++E +G + A+ + A Q CP + +W EA LE + +A
Sbjct 378 WIAAARLEELAGKLQAARELIATGCQQCPKSEDVWLEAARLE-------KPANAKAVLAK 430
Query 175 DVNVIHAVARL----FWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELE 222
V+V+ RL + R+K + + ++ + +++ P+ W ++ E E
Sbjct 431 AVSVLPHSVRLWFDAYAREKDLDQRKRVLRKALEFIPNSVRLWKEAVSLEEE 482
> dre:323855 c20orf14, fc12b02, prpf6, wu:fa05f07, wu:fc12b02,
zgc:65913; c20orf14 homolog (H. sapiens); K12855 pre-mRNA-processing
factor 6
Length=944
Score = 228 bits (580), Expect = 2e-59, Method: Compositional matrix adjust.
Identities = 112/257 (43%), Positives = 167/257 (64%), Gaps = 3/257 (1%)
Query 1 RQNINTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDL 60
R + TA V+M+S++LE LG+ +AA L EA+K ++ F KLWM+RGQ+ E+ +
Sbjct 672 RSSAPTARVFMKSVRLEWVLGNIEAAHELCTEALKHYEDFPKLWMMRGQIEEQ---SESI 728
Query 61 ARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQ 120
RA + + +G CP S+ LWL E RARA+LEKA+L+ P SPELWL V+
Sbjct 729 DRAREAYNQGLKKCPHSMSLWLLLSRLEEKVGQLTRARAILEKARLKNPQSPELWLESVR 788
Query 121 IEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCENDVNVIH 180
+E +G + +A + AKA+Q CPN+GI+W+EA+FLE + ++VDAL KCE+D +V+
Sbjct 789 LEYRAGLKNIANTLMAKALQECPNSGILWSEAVFLEARPQRKTKSVDALKKCEHDPHVLL 848
Query 181 AVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELENGGEKECIDIINRCALAQ 240
AVA+LFW ++KI KAR+W R+V ++P LGDAW + FEL++G E++ ++ RC A+
Sbjct 849 AVAKLFWSERKITKAREWFLRTVKIEPDLGDAWGFFYKFELQHGTEEQQHEVKKRCENAE 908
Query 241 PNRGLAWNKTTKQVRCW 257
P G W +K + W
Sbjct 909 PRHGELWCAESKHILNW 925
Score = 56.6 bits (135), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 57/250 (22%), Positives = 111/250 (44%), Gaps = 16/250 (6%)
Query 8 NVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQ---LHEEHPTQRD-LARA 63
+VW+++ E+ G ++ AL AV LW++ + L + P R LA A
Sbjct 578 SVWLRAAYFEKNNGTRESLEALLQRAVAHCPKAEVLWLMGAKSKWLAGDVPAARSILALA 637
Query 64 IDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEI 123
P S +WL V E + RAR +L KA+ P + +++ V++E
Sbjct 638 FQ-------ANPNSEEIWLAAVKLESENNEYERARRLLAKARSSAPTA-RVFMKSVRLEW 689
Query 124 ESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDA----LTKCENDVNVI 179
GN E A + +A++ + +W +EE++ RA +A L KC + +++
Sbjct 690 VLGNIEAAHELCTEALKHYEDFPKLWMMRGQIEEQSESIDRAREAYNQGLKKCPHSMSLW 749
Query 180 HAVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELENGGEKECIDIINRCALA 239
++RL + ++ +AR + ++ +P + W + E G + ++ +
Sbjct 750 LLLSRLEEKVGQLTRARAILEKARLKNPQSPELWLESVRLEYRAGLKNIANTLMAKALQE 809
Query 240 QPNRGLAWNK 249
PN G+ W++
Sbjct 810 CPNSGILWSE 819
Score = 50.8 bits (120), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 35/142 (24%), Positives = 67/142 (47%), Gaps = 14/142 (9%)
Query 6 TANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAID 65
+ +++++ +LE D A + +A++ +LW +L E + L+RA++
Sbjct 375 SVRIYIRAAELET---DIRAKKRVLRKALENVSKSVRLWKTAVELEEPEDARIMLSRAVE 431
Query 66 VFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIES 125
CCP SV LWL E + AR VL KA+ +P +W+ ++E +
Sbjct 432 -------CCPTSVELWLALARLE----TYENARRVLNKARENIPTDRHIWITAAKLEEAN 480
Query 126 GNREVAQHVAAKAIQACPNAGI 147
GN ++ + + +AI + G+
Sbjct 481 GNTQMVEKIIDRAITSLRANGV 502
Score = 45.8 bits (107), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 37/132 (28%), Positives = 59/132 (44%), Gaps = 11/132 (8%)
Query 61 ARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQ 120
A A+ VF P +WL E++ A+L++A P + LWL +
Sbjct 567 AHALQVF-------PSKKSVWLRAAYFEKNNGTRESLEALLQRAVAHCPKAEVLWLMGAK 619
Query 121 IEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCENDVNVIH 180
+ +G+ A+ + A A QA PN+ IW A+ LE EN + RA L K +
Sbjct 620 SKWLAGDVPAARSILALAFQANPNSEEIWLAAVKLESENNEYERARRLLAKARSSA---- 675
Query 181 AVARLFWRDKKI 192
AR+F + ++
Sbjct 676 PTARVFMKSVRL 687
Score = 42.4 bits (98), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 49/206 (23%), Positives = 88/206 (42%), Gaps = 14/206 (6%)
Query 44 WMVRGQLHEEHPTQRDLARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEK 103
W+ +L E L A ++ +GT CP+S +WL R Q + A+AV+ +
Sbjct 315 WIASARLEE---VTGKLQVARNLIMKGTEMCPKSEDVWL---EAARLQPG-DTAKAVVAQ 367
Query 104 AKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTH 163
A +P S +++ ++E + + V KA++ + +W A+ LEE
Sbjct 368 AVRHLPQSVRIYIRAAELETDI---RAKKRVLRKALENVSKSVRLWKTAVELEEPEDARI 424
Query 164 RAVDALTKCENDVNVIHAVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELEN 223
A+ C V + A+ARL + AR+ +N++ P+ W + E N
Sbjct 425 MLSRAVECCPTSVELWLALARL----ETYENARRVLNKARENIPTDRHIWITAAKLEEAN 480
Query 224 GGEKECIDIINRCALAQPNRGLAWNK 249
G + II+R + G+ N+
Sbjct 481 GNTQMVEKIIDRAITSLRANGVEINR 506
Score = 38.5 bits (88), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 18/61 (29%), Positives = 35/61 (57%), Gaps = 0/61 (0%)
Query 96 RARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFL 155
+AR +L+ + P+ P W+A ++E +G +VA+++ K + CP + +W EA L
Sbjct 296 KARLLLKSVRETNPHHPPAWIASARLEEVTGKLQVARNLIMKGTEMCPKSEDVWLEAARL 355
Query 156 E 156
+
Sbjct 356 Q 356
Score = 33.9 bits (76), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 26/53 (49%), Gaps = 0/53 (0%)
Query 97 ARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQACPNAGIIW 149
ARA+ A P+ +WL E +G RE + + +A+ CP A ++W
Sbjct 562 ARAIYAHALQVFPSKKSVWLRAAYFEKNNGTRESLEALLQRAVAHCPKAEVLW 614
> xla:447198 prpf6, MGC80263; PRP6 pre-mRNA processing factor
6 homolog; K12855 pre-mRNA-processing factor 6
Length=948
Score = 226 bits (576), Expect = 6e-59, Method: Compositional matrix adjust.
Identities = 113/257 (43%), Positives = 170/257 (66%), Gaps = 3/257 (1%)
Query 1 RQNINTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDL 60
R + TA V+M+S++LE LG+ +AA L +EA++ ++ F KLWM++GQ+ EE Q +
Sbjct 676 RSSAPTARVFMKSVKLEWVLGNIEAAQDLCEEALRHYEDFPKLWMMKGQI-EEQMEQTEK 734
Query 61 ARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQ 120
AR D + +G C S LWL E RARA+LEK++L+ P +PELWL V+
Sbjct 735 AR--DAYNQGLKKCIHSTSLWLLLSRLEEKVGQLTRARAILEKSRLKNPKTPELWLESVR 792
Query 121 IEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCENDVNVIH 180
+E +G + +A + AKA+Q CPN+GI+WAEA+FLE + ++VDAL KCE+D +V+
Sbjct 793 LEFRAGLKNIANTLMAKALQECPNSGILWAEAVFLEARPQRKTKSVDALKKCEHDPHVLL 852
Query 181 AVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELENGGEKECIDIINRCALAQ 240
AVA+LFW ++KI KAR+W +R+V +D LGDAWA++ FEL++G E++ +I RC A+
Sbjct 853 AVAKLFWSERKITKAREWFHRTVKIDSDLGDAWATFYKFELQHGTEEQQEEIRKRCENAE 912
Query 241 PNRGLAWNKTTKQVRCW 257
P G W +K ++ W
Sbjct 913 PRHGELWCAVSKDIKNW 929
Score = 63.2 bits (152), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 61/248 (24%), Positives = 110/248 (44%), Gaps = 16/248 (6%)
Query 8 NVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQ---LHEEHPTQRD-LARA 63
+VW+++ E+ G ++ AL AV LW++ + L + P R LA A
Sbjct 582 SVWLRAAYFEKNHGTRESLEALLQRAVAHCPKAEVLWLMGAKSKWLAGDVPAARSILALA 641
Query 64 IDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEI 123
P S +WL V E + RAR +L KA+ P + +++ V++E
Sbjct 642 FQ-------ANPNSEEIWLAAVKLESENNEYERARRLLAKARSSAPTA-RVFMKSVKLEW 693
Query 124 ESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDA----LTKCENDVNVI 179
GN E AQ + +A++ + +W +EE+ QT +A DA L KC + ++
Sbjct 694 VLGNIEAAQDLCEEALRHYEDFPKLWMMKGQIEEQMEQTEKARDAYNQGLKKCIHSTSLW 753
Query 180 HAVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELENGGEKECIDIINRCALA 239
++RL + ++ +AR + +S +P + W + E G + ++ +
Sbjct 754 LLLSRLEEKVGQLTRARAILEKSRLKNPKTPELWLESVRLEFRAGLKNIANTLMAKALQE 813
Query 240 QPNRGLAW 247
PN G+ W
Sbjct 814 CPNSGILW 821
Score = 49.7 bits (117), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 56/209 (26%), Positives = 97/209 (46%), Gaps = 20/209 (9%)
Query 44 WMVRGQLHEEHPTQRDLARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEK 103
W+ +L E L A ++ +GT CP+S +WL R Q + A+AV+ +
Sbjct 319 WIASARLEE---VTGKLQVARNLIMKGTEMCPKSEDVWL---EAARLQPG-DTAKAVVAQ 371
Query 104 AKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEE-ENAQT 162
A +P S +++ ++E + + V KA++ PN+ +W A+ LEE E+A+
Sbjct 372 AVRHLPQSVRIYIRAAELET---DLRAKKRVLRKALEHVPNSVRLWKAAVELEEPEDARI 428
Query 163 --HRAVDALTKCENDVNVIHAVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFE 220
RAV+ C +V + A+ARL + ARK +N++ P+ W + E
Sbjct 429 MLSRAVEC---CPTNVELWLALARL----ETYENARKVLNKARENIPTDRHIWITAAKLE 481
Query 221 LENGGEKECIDIINRCALAQPNRGLAWNK 249
NG + II+R + G+ N+
Sbjct 482 EANGNTQMVEKIIDRAITSLRANGVEINR 510
Score = 49.7 bits (117), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 34/142 (23%), Positives = 67/142 (47%), Gaps = 14/142 (9%)
Query 6 TANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAID 65
+ +++++ +LE D A + +A++ +LW +L E + L+RA++
Sbjct 379 SVRIYIRAAELET---DLRAKKRVLRKALEHVPNSVRLWKAAVELEEPEDARIMLSRAVE 435
Query 66 VFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIES 125
CCP +V LWL E + AR VL KA+ +P +W+ ++E +
Sbjct 436 -------CCPTNVELWLALARLE----TYENARKVLNKARENIPTDRHIWITAAKLEEAN 484
Query 126 GNREVAQHVAAKAIQACPNAGI 147
GN ++ + + +AI + G+
Sbjct 485 GNTQMVEKIIDRAITSLRANGV 506
Score = 47.0 bits (110), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 38/132 (28%), Positives = 59/132 (44%), Gaps = 6/132 (4%)
Query 61 ARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQ 120
ARAI + P +WL E++ A+L++A P + LWL +
Sbjct 566 ARAI--YAHSLQVFPSKKSVWLRAAYFEKNHGTRESLEALLQRAVAHCPKAEVLWLMGAK 623
Query 121 IEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCENDVNVIH 180
+ +G+ A+ + A A QA PN+ IW A+ LE EN + RA L K +
Sbjct 624 SKWLAGDVPAARSILALAFQANPNSEEIWLAAVKLESENNEYERARRLLAKARSSA---- 679
Query 181 AVARLFWRDKKI 192
AR+F + K+
Sbjct 680 PTARVFMKSVKL 691
Score = 38.5 bits (88), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 18/61 (29%), Positives = 35/61 (57%), Gaps = 0/61 (0%)
Query 96 RARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFL 155
+AR +L+ + P+ P W+A ++E +G +VA+++ K + CP + +W EA L
Sbjct 300 KARLLLKSVRETNPHHPPAWIASARLEEVTGKLQVARNLIMKGTEMCPKSEDVWLEAARL 359
Query 156 E 156
+
Sbjct 360 Q 360
> mmu:68879 Prpf6, 1190003A07Rik, 2610031L17Rik, ANT-1, MGC11655,
MGC36967, MGC38351, U5-102K; PRP6 pre-mRNA splicing factor
6 homolog (yeast); K12855 pre-mRNA-processing factor 6
Length=941
Score = 223 bits (568), Expect = 5e-58, Method: Compositional matrix adjust.
Identities = 108/257 (42%), Positives = 167/257 (64%), Gaps = 3/257 (1%)
Query 1 RQNINTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDL 60
R + TA V+M+S++LE LG+ AA L +EA++ ++ F KLWM++GQ+ E+ +
Sbjct 669 RSSAPTARVFMKSVKLEWVLGNISAAQELCEEALRHYEDFPKLWMMKGQIEEQGEL---M 725
Query 61 ARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQ 120
+A + + +G CP S PLWL E RARA+LEK++L+ P +P LWL V+
Sbjct 726 EKAREAYNQGLKKCPHSTPLWLLLSRLEEKIGQLTRARAILEKSRLKNPKNPGLWLESVR 785
Query 121 IEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCENDVNVIH 180
+E +G + +A + AKA+Q CPN+GI+W+EA+FLE + ++VDAL KCE+D +V+
Sbjct 786 LEYRAGLKNIANTLMAKALQECPNSGILWSEAVFLEARPQRKTKSVDALKKCEHDPHVLL 845
Query 181 AVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELENGGEKECIDIINRCALAQ 240
AVA+LFW ++KI KAR+W +R+V +D LGDAWA + FEL++G E++ ++ RC A+
Sbjct 846 AVAKLFWSERKITKAREWFHRTVKIDSDLGDAWAFFYKFELQHGTEEQQEEVRKRCENAE 905
Query 241 PNRGLAWNKTTKQVRCW 257
P G W +K + W
Sbjct 906 PRHGELWCAVSKDITNW 922
Score = 53.5 bits (127), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 57/250 (22%), Positives = 107/250 (42%), Gaps = 16/250 (6%)
Query 8 NVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQ---LHEEHPTQRD-LARA 63
+VW+++ E+ G ++ AL AV LW++ + L + P R LA A
Sbjct 575 SVWLRAAYFEKNHGTRESLEALLQRAVAHCPKAEVLWLMGAKSKWLAGDVPAARSILALA 634
Query 64 IDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEI 123
P S +WL V E + RAR +L KA+ P + +++ V++E
Sbjct 635 FQ-------ANPNSEEIWLAAVKLESENNEYERARRLLAKARSSAPTA-RVFMKSVKLEW 686
Query 124 ESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDA----LTKCENDVNVI 179
GN AQ + +A++ + +W +EE+ +A +A L KC + +
Sbjct 687 VLGNISAAQELCEEALRHYEDFPKLWMMKGQIEEQGELMEKAREAYNQGLKKCPHSTPLW 746
Query 180 HAVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELENGGEKECIDIINRCALA 239
++RL + ++ +AR + +S +P W + E G + ++ +
Sbjct 747 LLLSRLEEKIGQLTRARAILEKSRLKNPKNPGLWLESVRLEYRAGLKNIANTLMAKALQE 806
Query 240 QPNRGLAWNK 249
PN G+ W++
Sbjct 807 CPNSGILWSE 816
Score = 50.8 bits (120), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 35/142 (24%), Positives = 67/142 (47%), Gaps = 14/142 (9%)
Query 6 TANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAID 65
+ +++++ +LE D A + +A++ +LW +L E + L+RA++
Sbjct 372 SVRIYIRAAELET---DIRAKKRVLRKALEHVPNSVRLWKAAVELEEPEDARIMLSRAVE 428
Query 66 VFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIES 125
CCP SV LWL E + AR VL KA+ +P +W+ ++E +
Sbjct 429 -------CCPTSVELWLALARLE----TYENARKVLNKARENIPTDRHIWITAAKLEEAN 477
Query 126 GNREVAQHVAAKAIQACPNAGI 147
GN ++ + + +AI + G+
Sbjct 478 GNTQMVEKIIDRAITSLRANGV 499
Score = 49.3 bits (116), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 56/209 (26%), Positives = 96/209 (45%), Gaps = 20/209 (9%)
Query 44 WMVRGQLHEEHPTQRDLARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEK 103
W+ +L E L A ++ +GT CP+S +WL R Q + A+AV+ +
Sbjct 312 WIASARLEE---VTGKLQVARNLIMKGTEMCPKSEDVWL---EAARLQPG-DTAKAVVAQ 364
Query 104 AKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEE-ENAQT 162
A +P S +++ ++E + + V KA++ PN+ +W A+ LEE E+A+
Sbjct 365 AVRHLPQSVRIYIRAAELETDI---RAKKRVLRKALEHVPNSVRLWKAAVELEEPEDARI 421
Query 163 --HRAVDALTKCENDVNVIHAVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFE 220
RAV+ C V + A+ARL + ARK +N++ P+ W + E
Sbjct 422 MLSRAVEC---CPTSVELWLALARL----ETYENARKVLNKARENIPTDRHIWITAAKLE 474
Query 221 LENGGEKECIDIINRCALAQPNRGLAWNK 249
NG + II+R + G+ N+
Sbjct 475 EANGNTQMVEKIIDRAITSLRANGVEINR 503
Score = 46.6 bits (109), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 39/132 (29%), Positives = 60/132 (45%), Gaps = 6/132 (4%)
Query 61 ARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQ 120
ARAI + +SV WL E++ A+L++A P + LWL +
Sbjct 559 ARAIYAYALQVFPSKKSV--WLRAAYFEKNHGTRESLEALLQRAVAHCPKAEVLWLMGAK 616
Query 121 IEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCENDVNVIH 180
+ +G+ A+ + A A QA PN+ IW A+ LE EN + RA L K +
Sbjct 617 SKWLAGDVPAARSILALAFQANPNSEEIWLAAVKLESENNEYERARRLLAKARSSA---- 672
Query 181 AVARLFWRDKKI 192
AR+F + K+
Sbjct 673 PTARVFMKSVKL 684
Score = 38.5 bits (88), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 18/61 (29%), Positives = 35/61 (57%), Gaps = 0/61 (0%)
Query 96 RARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFL 155
+AR +L+ + P+ P W+A ++E +G +VA+++ K + CP + +W EA L
Sbjct 293 KARLLLKSVRETNPHHPPAWIASARLEEVTGKLQVARNLIMKGTEMCPKSEDVWLEAARL 352
Query 156 E 156
+
Sbjct 353 Q 353
> hsa:24148 PRPF6, ANT-1, C20orf14, Prp6, TOM, U5-102K, hPrp6;
PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae);
K12855 pre-mRNA-processing factor 6
Length=941
Score = 217 bits (552), Expect = 4e-56, Method: Compositional matrix adjust.
Identities = 107/257 (41%), Positives = 165/257 (64%), Gaps = 3/257 (1%)
Query 1 RQNINTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDL 60
R + TA V+M+S++LE + AA L +EA++ ++ F KLWM++GQ+ E+ + +
Sbjct 669 RSSAPTARVFMKSVKLEWVQDNIRAAQDLCEEALRHYEDFPKLWMMKGQIEEQ---KEMM 725
Query 61 ARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQ 120
+A + + +G CP S PLWL E RARA+LEK++L+ P +P LWL V+
Sbjct 726 EKAREAYNQGLKKCPHSTPLWLLLSRLEEKIGQLTRARAILEKSRLKNPKNPGLWLESVR 785
Query 121 IEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCENDVNVIH 180
+E +G + +A + AKA+Q CPN+GI+W+EAIFLE + ++VDAL KCE+D +V+
Sbjct 786 LEYRAGLKNIANTLMAKALQECPNSGILWSEAIFLEARPQRRTKSVDALKKCEHDPHVLL 845
Query 181 AVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELENGGEKECIDIINRCALAQ 240
AVA+LFW +KI KAR+W +R+V +D LGDAWA + FEL++G E++ ++ RC A+
Sbjct 846 AVAKLFWSQRKITKAREWFHRTVKIDSDLGDAWAFFYKFELQHGTEEQQEEVRKRCESAE 905
Query 241 PNRGLAWNKTTKQVRCW 257
P G W +K + W
Sbjct 906 PRHGELWCAVSKDIANW 922
Score = 50.8 bits (120), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 35/142 (24%), Positives = 67/142 (47%), Gaps = 14/142 (9%)
Query 6 TANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAID 65
+ +++++ +LE D A + +A++ +LW +L E + L+RA++
Sbjct 372 SVRIYIRAAELE---TDIRAKKRVLRKALEHVPNSVRLWKAAVELEEPEDARIMLSRAVE 428
Query 66 VFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIES 125
CCP SV LWL E + AR VL KA+ +P +W+ ++E +
Sbjct 429 -------CCPTSVELWLALARLE----TYENARKVLNKARENIPTDRHIWITAAKLEEAN 477
Query 126 GNREVAQHVAAKAIQACPNAGI 147
GN ++ + + +AI + G+
Sbjct 478 GNTQMVEKIIDRAITSLRANGV 499
Score = 49.3 bits (116), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 52/206 (25%), Positives = 90/206 (43%), Gaps = 14/206 (6%)
Query 44 WMVRGQLHEEHPTQRDLARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEK 103
W+ +L E L A ++ +GT CP+S +WL R Q + A+AV+ +
Sbjct 312 WIASARLEE---VTGKLQVARNLIMKGTEMCPKSEDVWL---EAARLQPG-DTAKAVVAQ 364
Query 104 AKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTH 163
A +P S +++ ++E + + V KA++ PN+ +W A+ LEE
Sbjct 365 AVRHLPQSVRIYIRAAELETDI---RAKKRVLRKALEHVPNSVRLWKAAVELEEPEDARI 421
Query 164 RAVDALTKCENDVNVIHAVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELEN 223
A+ C V + A+ARL + ARK +N++ P+ W + E N
Sbjct 422 MLSRAVECCPTSVELWLALARL----ETYENARKVLNKARENIPTDRHIWITAAKLEEAN 477
Query 224 GGEKECIDIINRCALAQPNRGLAWNK 249
G + II+R + G+ N+
Sbjct 478 GNTQMVEKIIDRAITSLRANGVEINR 503
Score = 47.0 bits (110), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 38/132 (28%), Positives = 59/132 (44%), Gaps = 6/132 (4%)
Query 61 ARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQ 120
ARAI + P +WL E++ A+L++A P + LWL +
Sbjct 559 ARAIYAY--ALQVFPSKKSVWLRAAYFEKNHGTRESLEALLQRAVAHCPKAEVLWLMGAK 616
Query 121 IEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCENDVNVIH 180
+ +G+ A+ + A A QA PN+ IW A+ LE EN + RA L K +
Sbjct 617 SKWLAGDVPAARSILALAFQANPNSEEIWLAAVKLESENDEYERARRLLAKARSSA---- 672
Query 181 AVARLFWRDKKI 192
AR+F + K+
Sbjct 673 PTARVFMKSVKL 684
Score = 38.5 bits (88), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 18/61 (29%), Positives = 35/61 (57%), Gaps = 0/61 (0%)
Query 96 RARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFL 155
+AR +L+ + P+ P W+A ++E +G +VA+++ K + CP + +W EA L
Sbjct 293 KARLLLKSVRETNPHHPPAWIASARLEEVTGKLQVARNLIMKGTEMCPKSEDVWLEAARL 352
Query 156 E 156
+
Sbjct 353 Q 353
> hsa:653889 pre-mRNA-processing factor 6-like
Length=406
Score = 216 bits (550), Expect = 6e-56, Method: Compositional matrix adjust.
Identities = 107/257 (41%), Positives = 164/257 (63%), Gaps = 3/257 (1%)
Query 1 RQNINTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDL 60
R + TA V+M+S++LE + AA L +EA++ ++ F KLWM++GQ+ E+ +
Sbjct 134 RSSAPTARVFMKSVKLEWVQDNIRAAQDLCEEALRHYEDFPKLWMMKGQIEEQKEM---M 190
Query 61 ARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQ 120
+A + + +G CP S PLWL E RARA+LEK++L+ P +P LWL V+
Sbjct 191 EKAREAYNQGLKKCPHSTPLWLLLSRLEEKIGQLTRARAILEKSRLKNPKNPGLWLESVR 250
Query 121 IEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCENDVNVIH 180
+E +G + +A + AKA+Q CPN+GI+W+EAIFLE + ++VDAL KCE+D +V+
Sbjct 251 LEYRAGLKNIANTLMAKALQECPNSGILWSEAIFLEARPQRRTKSVDALKKCEHDPHVLL 310
Query 181 AVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELENGGEKECIDIINRCALAQ 240
AVA+LFW +KI KAR+W +R+V +D LGDAWA + FEL++G E++ ++ RC A+
Sbjct 311 AVAKLFWSQRKITKAREWFHRTVKIDSDLGDAWAFFYKFELQHGTEEQQEEVRKRCESAE 370
Query 241 PNRGLAWNKTTKQVRCW 257
P G W +K + W
Sbjct 371 PRHGELWCAVSKDIANW 387
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 52/246 (21%), Positives = 103/246 (41%), Gaps = 8/246 (3%)
Query 8 NVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAIDVF 67
+VW+++ E+ G ++ AL AV LW++ + D+ A +
Sbjct 40 SVWLRAAYFEKNHGTRESLEALLQRAVAHCPKAEVLWLMGAK---SKWLAGDVPAARSIL 96
Query 68 REGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGN 127
P S +WL V E + RAR +L KA+ P + +++ V++E N
Sbjct 97 ALAFQANPNSEEIWLAAVKLESENDEYERARRLLAKARSSAPTA-RVFMKSVKLEWVQDN 155
Query 128 REVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDA----LTKCENDVNVIHAVA 183
AQ + +A++ + +W +EE+ +A +A L KC + + ++
Sbjct 156 IRAAQDLCEEALRHYEDFPKLWMMKGQIEEQKEMMEKAREAYNQGLKKCPHSTPLWLLLS 215
Query 184 RLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELENGGEKECIDIINRCALAQPNR 243
RL + ++ +AR + +S +P W + E G + ++ + PN
Sbjct 216 RLEEKIGQLTRARAILEKSRLKNPKNPGLWLESVRLEYRAGLKNIANTLMAKALQECPNS 275
Query 244 GLAWNK 249
G+ W++
Sbjct 276 GILWSE 281
Score = 46.6 bits (109), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 38/132 (28%), Positives = 59/132 (44%), Gaps = 6/132 (4%)
Query 61 ARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQ 120
ARAI + P +WL E++ A+L++A P + LWL +
Sbjct 24 ARAIYAY--ALQVFPSKKSVWLRAAYFEKNHGTRESLEALLQRAVAHCPKAEVLWLMGAK 81
Query 121 IEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCENDVNVIH 180
+ +G+ A+ + A A QA PN+ IW A+ LE EN + RA L K +
Sbjct 82 SKWLAGDVPAARSILALAFQANPNSEEIWLAAVKLESENDEYERARRLLAKARSSA---- 137
Query 181 AVARLFWRDKKI 192
AR+F + K+
Sbjct 138 PTARVFMKSVKL 149
> ath:AT4G03430 EMB2770 (EMBRYO DEFECTIVE 2770); RNA splicing
factor, transesterification mechanism; K12855 pre-mRNA-processing
factor 6
Length=1029
Score = 199 bits (507), Expect = 7e-51, Method: Compositional matrix adjust.
Identities = 96/254 (37%), Positives = 151/254 (59%), Gaps = 3/254 (1%)
Query 1 RQNINTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDL 60
R+ T VWM+S +ER+LG+ + L +E +KQ TF KLW++ GQL E + L
Sbjct 754 RERGGTERVWMKSAIVERELGNVEEERRLLNEGLKQFPTFFKLWLMLGQLEERF---KHL 810
Query 61 ARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQ 120
+A + G CP +PLWL + E N+ARA+L A+ + P ELWLA ++
Sbjct 811 EQARKAYDTGLKHCPHCIPLWLSLADLEEKVNGLNKARAILTTARKKNPGGAELWLAAIR 870
Query 121 IEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCENDVNVIH 180
E+ N+ A+H+ +KA+Q CP +GI+WA I + + +++DA+ KC+ D +V
Sbjct 871 AELRHDNKREAEHLMSKALQDCPKSGILWAADIEMAPRPRRKTKSIDAMKKCDRDPHVTI 930
Query 181 AVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELENGGEKECIDIINRCALAQ 240
AVA+LFW+DKK+ KAR W R+VT+ P +GD WA + FEL++G +++ +++ +C +
Sbjct 931 AVAKLFWQDKKVEKARAWFERAVTVGPDIGDFWALFYKFELQHGSDEDRKEVVAKCVACE 990
Query 241 PNRGLAWNKTTKQV 254
P G W +K V
Sbjct 991 PKHGEKWQAISKAV 1004
Score = 58.9 bits (141), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 57/248 (22%), Positives = 108/248 (43%), Gaps = 8/248 (3%)
Query 4 INTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARA 63
+ ++W+++ QLE+ G ++ AL +AV LW++ + E D+ A
Sbjct 656 LTKKSIWLKAAQLEKSHGSRESLDALLRKAVTYVPQAEVLWLMGAK---EKWLAGDVPAA 712
Query 64 IDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEI 123
+ +E P S +WL E K RAR +L KA+ R + +W+ +E
Sbjct 713 RAILQEAYAAIPNSEEIWLAAFKLEFENKEPERARMLLAKARER-GGTERVWMKSAIVER 771
Query 124 ESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENA---QTHRAVD-ALTKCENDVNVI 179
E GN E + + + ++ P +W LEE Q +A D L C + + +
Sbjct 772 ELGNVEEERRLLNEGLKQFPTFFKLWLMLGQLEERFKHLEQARKAYDTGLKHCPHCIPLW 831
Query 180 HAVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELENGGEKECIDIINRCALA 239
++A L + + KAR + + +P + W + + EL + ++E ++++
Sbjct 832 LSLADLEEKVNGLNKARAILTTARKKNPGGAELWLAAIRAELRHDNKREAEHLMSKALQD 891
Query 240 QPNRGLAW 247
P G+ W
Sbjct 892 CPKSGILW 899
Score = 41.2 bits (95), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 34/157 (21%), Positives = 69/157 (43%), Gaps = 17/157 (10%)
Query 5 NTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAI 64
N+ +W+++ +LE + + + E + +LW +L E + L RA+
Sbjct 449 NSVKLWLEAAKLEHDVENKSRVLRKGLEHIPDS---VRLWKAVVELANEEDARILLHRAV 505
Query 65 DVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIE 124
+ CCP + LW+ E + ++ VL KA+ ++P P +W+ ++E
Sbjct 506 E-------CCPLHLELWVALARLE----TYAESKKVLNKAREKLPKEPAIWITAAKLEEA 554
Query 125 SGNREVAQHVAAKAIQACPNAGI--IWAEAIFLEEEN 159
+G + A A + + GI + E + ++ EN
Sbjct 555 NGKLDEANDNTA-MVGKIIDRGIKTLQREGVVIDREN 590
Score = 33.9 bits (76), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 31/128 (24%), Positives = 54/128 (42%), Gaps = 5/128 (3%)
Query 59 DLARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLAL 118
D RA +++ T P++ W+ E AR +++ P + ++WL
Sbjct 368 DRNRARLLYKSLTQSNPKNPNGWIAAARVEEVDGKIKAARFQIQRGCEECPKNEDVWLEA 427
Query 119 VQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAV-DALTKCENDVN 177
++ N E A+ V AK ++ PN+ +W EA LE + R + L + V
Sbjct 428 CRL----ANPEDAKGVIAKGVKLIPNSVKLWLEAAKLEHDVENKSRVLRKGLEHIPDSVR 483
Query 178 VIHAVARL 185
+ AV L
Sbjct 484 LWKAVVEL 491
> tpv:TP01_0797 hypothetical protein; K12855 pre-mRNA-processing
factor 6
Length=1032
Score = 192 bits (488), Expect = 1e-48, Method: Compositional matrix adjust.
Identities = 113/324 (34%), Positives = 172/324 (53%), Gaps = 64/324 (19%)
Query 1 RQNINTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLH-EEHPTQRD 59
R NT VWM+S+QLERQL +Y+ A+ L D+A++ H F KLWM+ GQL E+HP +D
Sbjct 674 RTRCNTPKVWMKSVQLERQLKNYEKALELVDKALEIHPYFDKLWMISGQLKLEKHP--KD 731
Query 60 LARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRV----------- 108
+ A +++G CP SV LWL + + K + +ARA++E AK ++
Sbjct 732 VEGATLTYKQGVETCPWSVNLWLLSIELQIELKEFAKARALVETAKNKIRTILGSNIKKN 791
Query 109 ----------------------------PNSPE------------LWLALVQIEIESGNR 128
P+S + +WL V+IE+E+G
Sbjct 792 TDITKVQTKVLSNSELSRMAKLSMDSDDPDSVKEMIENIISQCDLIWLKGVEIEMETGVH 851
Query 129 EVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCENDVNVIHAVARLFWR 188
E A +KA+Q P++G++WA +IFLEE NAQ +A +AL + +N +++ A A++FW
Sbjct 852 ENAHFAMSKALQELPDSGLLWARSIFLEEPNAQKTKAAEALKRNQNSPHIVLAAAKIFWN 911
Query 189 DKKIAKARKWMNRSVTLDPSLGDAW----------ASYLAFELENGGEKECIDIINRCAL 238
K I KAR+W +TLD S G +W +++AFEL+ G E+ IN+
Sbjct 912 CKMIDKARRWFQTCITLDESNGISWGIFVKFMNVLGTFIAFELDCGTEESMKQAINKFIE 971
Query 239 AQPNRGLAWNKTTKQVRCWSLSLP 262
A+PNRG W + TK+V W++ LP
Sbjct 972 AEPNRGYEWCRVTKKVENWNIPLP 995
Score = 57.8 bits (138), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 44/179 (24%), Positives = 75/179 (41%), Gaps = 38/179 (21%)
Query 5 NTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAI 64
N +VW++ + + G Y+ A AL A++ +T + LW+ +L +H T +
Sbjct 543 NRKSVWLEDGETFVEHGSYECARALYKNALEYMKTRSSLWLALVELESKHGTPDKVE--- 599
Query 65 DVFREGTVCCPRSVPLWLCW----------------------------------VNCERS 90
D R CP S LWL + ++
Sbjct 600 DHLRSAVSYCPNSEILWLMYAKHKWVEGDVESSRDILSRALTMNENNEAISLAAAKLDKE 659
Query 91 QKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQACPNAGIIW 149
++RAR +LEKA+ R N+P++W+ VQ+E + N E A + KA++ P +W
Sbjct 660 THEYDRARKLLEKARTRC-NTPKVWMKSVQLERQLKNYEKALELVDKALEIHPYFDKLW 717
Score = 41.2 bits (95), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 32/114 (28%), Positives = 58/114 (50%), Gaps = 8/114 (7%)
Query 97 ARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLE 156
ARA+ + A + LWLALV++E + G + + A+ CPN+ I+W ++ +
Sbjct 564 ARALYKNALEYMKTRSSLWLALVELESKHGTPDKVEDHLRSAVSYCPNSEILW--LMYAK 621
Query 157 ----EENAQTHRAV--DALTKCENDVNVIHAVARLFWRDKKIAKARKWMNRSVT 204
E + ++ R + ALT EN+ + A A+L + +ARK + ++ T
Sbjct 622 HKWVEGDVESSRDILSRALTMNENNEAISLAAAKLDKETHEYDRARKLLEKART 675
Score = 40.8 bits (94), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 52/202 (25%), Positives = 76/202 (37%), Gaps = 43/202 (21%)
Query 3 NINTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLAR 62
N A W+ + ++E G +AA + + + +W+ +L + + LA+
Sbjct 310 NQKHAQGWIAAARMEELAGKIEAAREIIAQGCENCPDKEDVWLEAARLEKPEYAKSILAK 369
Query 63 AIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIE 122
AI + P SV LWL + E S N R VL KA +PNS LW + +E
Sbjct 370 AIKII-------PTSVKLWLEAADKETSNDNRRR---VLRKALEFIPNSIRLWKEAISLE 419
Query 123 IESG--------------------------NREVAQHVAAKAIQACPNAGIIWAEAIFLE 156
E+ E AQ V +A + P IW A LE
Sbjct 420 NETNAYILLKRAVECVPESLDMWLALARLCPYEEAQKVLNEARKKLPTNVDIWITAAKLE 479
Query 157 EENAQTH-------RAVDALTK 171
E N RA+D L+K
Sbjct 480 ESNNNYEMVERIIVRAIDNLSK 501
Score = 40.4 bits (93), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 32/166 (19%), Positives = 70/166 (42%), Gaps = 34/166 (20%)
Query 8 NVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLAR----- 62
+VW+++ +LE+ + A ++ +A+K T KLW+ + +R + R
Sbjct 349 DVWLEAARLEKP----EYAKSILAKAIKIIPTSVKLWLEAADKETSNDNRRRVLRKALEF 404
Query 63 -------------------AIDVFREGTVCCPRSVPLWLCWVN-CERSQKNWNRARAVLE 102
A + + C P S+ +WL C + A+ VL
Sbjct 405 IPNSIRLWKEAISLENETNAYILLKRAVECVPESLDMWLALARLCP-----YEEAQKVLN 459
Query 103 KAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQACPNAGII 148
+A+ ++P + ++W+ ++E + N E+ + + +AI G++
Sbjct 460 EARKKLPTNVDIWITAAKLEESNNNYEMVERIIVRAIDNLSKKGVV 505
Score = 36.2 bits (82), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 43/184 (23%), Positives = 76/184 (41%), Gaps = 8/184 (4%)
Query 43 LWMVRGQLHEEHPTQRDLARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLE 102
+W+ G+ EH + + ARA+ ++ LWL V E ++ L
Sbjct 547 VWLEDGETFVEHGSY-ECARAL--YKNALEYMKTRSSLWLALVELESKHGTPDKVEDHLR 603
Query 103 KAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQT 162
A PNS LWL + + G+ E ++ + ++A+ N I A L++E +
Sbjct 604 SAVSYCPNSEILWLMYAKHKWVEGDVESSRDILSRALTMNENNEAISLAAAKLDKETHEY 663
Query 163 HRAVDAL----TKCENDVNVIHAVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLA 218
RA L T+C N V +L + K KA + +++++ + P W
Sbjct 664 DRARKLLEKARTRC-NTPKVWMKSVQLERQLKNYEKALELVDKALEIHPYFDKLWMISGQ 722
Query 219 FELE 222
+LE
Sbjct 723 LKLE 726
Score = 35.4 bits (80), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 24/98 (24%), Positives = 44/98 (44%), Gaps = 0/98 (0%)
Query 60 LARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALV 119
L + D TV P+ L + E + + +AR +L+ + W+A
Sbjct 262 LDKVTDNLSGQTVVDPKGYLTDLNSMKTEFEEADVQKARTLLKSLISTNQKHAQGWIAAA 321
Query 120 QIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEE 157
++E +G E A+ + A+ + CP+ +W EA LE+
Sbjct 322 RMEELAGKIEAAREIIAQGCENCPDKEDVWLEAARLEK 359
> cel:Y59A8B.6 hypothetical protein; K12855 pre-mRNA-processing
factor 6
Length=968
Score = 177 bits (449), Expect = 3e-44, Method: Compositional matrix adjust.
Identities = 94/257 (36%), Positives = 152/257 (59%), Gaps = 3/257 (1%)
Query 1 RQNINTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDL 60
R +A VWM++ E LG+ + A L +E ++++ F K+++V GQ+ E+ D+
Sbjct 696 RAKAPSARVWMKNAHFEWCLGNVEEAKRLCEECIQKYDDFHKIYLVLGQVLEQ---MNDV 752
Query 61 ARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQ 120
A + +G CP +PLW+ V E +AR LEKA+LR P + +LWL V+
Sbjct 753 HGARLAYTQGIRKCPGVIPLWILLVRLEEKAGQIVKARVDLEKARLRNPKNDDLWLESVR 812
Query 121 IEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCENDVNVIH 180
E G E+A+ ++A+Q C +G +WAEAI++E + + +++DAL KCE++ +V+
Sbjct 813 FEQRVGCPEMAKERMSRALQECEGSGKLWAEAIWMEGPHGRRAKSIDALKKCEHNPHVLI 872
Query 181 AVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELENGGEKECIDIINRCALAQ 240
A ARLFW ++KI KAR+W R+V LDP GDA+A++LAFE +G E++ + +C ++
Sbjct 873 AAARLFWSERKIKKAREWFVRAVNLDPDNGDAFANFLAFEQIHGKEEDRKSVFKKCVTSE 932
Query 241 PNRGLAWNKTTKQVRCW 257
P G W +K W
Sbjct 933 PRYGDLWQSVSKDPINW 949
Score = 51.2 bits (121), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 34/137 (24%), Positives = 64/137 (46%), Gaps = 14/137 (10%)
Query 5 NTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAI 64
++ +W ++ LE+ L D + +A++Q + KLW +L + + L RA+
Sbjct 397 HSVRLWCKASDLEQDLKDKKKVLR---KALEQIPSSVKLWKAAVELEDPEDARILLTRAV 453
Query 65 DVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIE 124
+ CC S +WL E + AR VL KA+ +P +WL+ ++E
Sbjct 454 E-------CCSSSTEMWLALARLE----TYENARKVLNKAREHIPTDRHIWLSAARLEET 502
Query 125 SGNREVAQHVAAKAIQA 141
G +++ + AKA+ +
Sbjct 503 RGQKDMVDKIVAKAMSS 519
Score = 45.8 bits (107), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 44/153 (28%), Positives = 73/153 (47%), Gaps = 15/153 (9%)
Query 10 WMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAIDVFRE 69
W+ S LE Q G A E ++ + +LW+ +LH +L R+I
Sbjct 338 WVASAVLEEQAGKLQTARNFIMEGCEKIKNSEELWLHAIRLHPP-----ELGRSI--VAN 390
Query 70 GTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNRE 129
CP SV LW + E+ K+ + VL KA ++P+S +LW A V++E + E
Sbjct 391 AVRSCPHSVRLWCKASDLEQDLKD---KKKVLRKALEQIPSSVKLWKAAVELE----DPE 443
Query 130 VAQHVAAKAIQACPNAGIIWAEAIFLEE-ENAQ 161
A+ + +A++ C ++ +W LE ENA+
Sbjct 444 DARILLTRAVECCSSSTEMWLALARLETYENAR 476
Score = 41.6 bits (96), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 46/158 (29%), Positives = 68/158 (43%), Gaps = 20/158 (12%)
Query 5 NTANVWMQSIQLER-QLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARA 63
N+ +W+ +I+L +LG ++ AV+ +LW L ++DL
Sbjct 367 NSEELWLHAIRLHPPELGR-----SIVANAVRSCPHSVRLWCKASDL------EQDLKDK 415
Query 64 IDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEI 123
V R+ P SV LW V E + AR +L +A +S E+WLAL ++E
Sbjct 416 KKVLRKALEQIPSSVKLWKAAVELEDPED----ARILLTRAVECCSSSTEMWLALARLE- 470
Query 124 ESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQ 161
E A+ V KA + P IW A LEE Q
Sbjct 471 ---TYENARKVLNKAREHIPTDRHIWLSAARLEETRGQ 505
Score = 40.0 bits (92), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 57/228 (25%), Positives = 95/228 (41%), Gaps = 21/228 (9%)
Query 8 NVWMQSIQLERQLGDYD---AAIALADEAVKQHQTF----AKLWMVRGQLHEEHPTQRDL 60
++W +I ER+ G D A + A E V + + + AKL V ++ E T L
Sbjct 601 SIWDAAIHFEREHGSLDEHEAILLKACETVPEVEDYWLMLAKLRFVNKRVGEARDT---L 657
Query 61 ARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQ 120
A + + G +S +WL E ++ AR + KA+ + P S +W+
Sbjct 658 MSAFE--KHGH----QSEKIWLAATKIEIETDQFDTARGLFGKARAKAP-SARVWMKNAH 710
Query 121 IEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALT----KCENDV 176
E GN E A+ + + IQ + I+ + E+ H A A T KC +
Sbjct 711 FEWCLGNVEEAKRLCEECIQKYDDFHKIYLVLGQVLEQMNDVHGARLAYTQGIRKCPGVI 770
Query 177 NVIHAVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELENG 224
+ + RL + +I KAR + ++ +P D W + FE G
Sbjct 771 PLWILLVRLEEKAGQIVKARVDLEKARLRNPKNDDLWLESVRFEQRVG 818
Score = 40.0 bits (92), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 24/101 (23%), Positives = 49/101 (48%), Gaps = 4/101 (3%)
Query 58 RDLARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLA 117
+D+ +A + + PR P W+ E AR + + ++ NS ELWL
Sbjct 315 QDIKKARMLLKSVRETNPRHPPAWVASAVLEEQAGKLQTARNFIMEGCEKIKNSEELWLH 374
Query 118 LVQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEE 158
+++ E+ + + A A+++CP++ +W +A LE++
Sbjct 375 AIRLHPP----ELGRSIVANAVRSCPHSVRLWCKASDLEQD 411
Score = 30.8 bits (68), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 27/132 (20%), Positives = 59/132 (44%), Gaps = 5/132 (3%)
Query 92 KNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQACPNAGIIWAE 151
++ +AR +L+ + P P W+A +E ++G + A++ + + N+ +W
Sbjct 315 QDIKKARMLLKSVRETNPRHPPAWVASAVLEEQAGKLQTARNFIMEGCEKIKNSEELWLH 374
Query 152 AIFLEEENAQTHRAVDALTKCENDVNVIHAVARLFWRDKKIAKARKWMNRSVTLDPSLGD 211
AI L +A+ C + V + + L ++ + +K + +++ PS
Sbjct 375 AIRLHPPELGRSIVANAVRSCPHSVRLWCKASDL---EQDLKDKKKVLRKALEQIPSSVK 431
Query 212 AWASYLAFELEN 223
W + A ELE+
Sbjct 432 LWKA--AVELED 441
> cpv:cgd5_920 Pre-mRNA splicing factor Pro1/Prp6. HAT repeat
protein ; K12855 pre-mRNA-processing factor 6
Length=923
Score = 172 bits (437), Expect = 8e-43, Method: Composition-based stats.
Identities = 84/257 (32%), Positives = 143/257 (55%), Gaps = 1/257 (0%)
Query 1 RQNINTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDL 60
R N + +W++SI+LE +YD I E+VK++ + LW++ G ++ + R +
Sbjct 633 RTNSPSVQIWVESIKLENDQKNYDLCILYCSESVKEYPSSPNLWLLYGFIYRKAFPDR-I 691
Query 61 ARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQ 120
A+ ++ EG C S+ LW + +NW +AR L+ A+ + N PELW+ ++
Sbjct 692 NEALKIYEEGLNFCSDSIELWFSTIELLMLLQNWKKARTFLDLARSKNKNQPELWMQTIK 751
Query 121 IEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCENDVNVIH 180
+E +GN E + +KA++ CP +G+++AE+IF E++ Q + + AL +C ND V+
Sbjct 752 LEKNAGNNEFIPQILSKALKECPKSGLLYAESIFTEQKQKQKSKFLIALEQCGNDPYVLV 811
Query 181 AVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELENGGEKECIDIINRCALAQ 240
A+A FW++ K+RKW ++ +D +GD W Y+AFEL NG + D +N A
Sbjct 812 AIAISFWKENDFHKSRKWFKSALEIDNKIGDTWIHYIAFELLNGDFQSQRDALNDFINAT 871
Query 241 PNRGLAWNKTTKQVRCW 257
PN+G WN + W
Sbjct 872 PNKGFEWNNIRRTHFFW 888
Score = 46.6 bits (109), Expect = 9e-05, Method: Composition-based stats.
Identities = 54/241 (22%), Positives = 100/241 (41%), Gaps = 33/241 (13%)
Query 3 NINTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLAR 62
++NTA I+L L D A L V + + W+ + E L+
Sbjct 239 SLNTA-----GIKLNGDLSDIKKARLLLKSVVNTNPKHSPGWIAAARFEE---FVGRLSH 290
Query 63 AIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIE 122
A ++ +G CP++ +WL + + ++ ++ K+ +PNS ++W+
Sbjct 291 AREIIAKGCEMCPKNEDIWLEAIRLGKPEQ----IDKIIVKSIKFIPNSTKVWMV----- 341
Query 123 IESGNREV----AQHVAAKAIQACPNAGIIWAEAIFL---EEENAQTHRAVDALTKCEND 175
+ NRE + KA++ PN+ +W EAI L E E A +AV + + E
Sbjct 342 --AANRETNKNKKLLIIKKALEFIPNSIKLWKEAISLVDNESEKALLSKAVKCVPQSEE- 398
Query 176 VNVIHAVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELENGGEKECIDIINR 235
+ + R + A+K +N + + P+ W E +NG ++ I+ R
Sbjct 399 ------LWLRYARLSEYCDAQKILNEARKVLPTFPGIWVEAAKLEEQNGKVEKVELIVKR 452
Query 236 C 236
C
Sbjct 453 C 453
Score = 44.3 bits (103), Expect = 5e-04, Method: Composition-based stats.
Identities = 34/139 (24%), Positives = 57/139 (41%), Gaps = 6/139 (4%)
Query 97 ARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLE 156
ARA+ E + + W+ E + GN E HV K+++ CP+ I+W +A +
Sbjct 522 ARAMFESSADMFKSKEYFWIKWANFEEKYGNFEKVDHVLQKSLKNCPDKQILWLKAAQNQ 581
Query 157 EENAQTHRAVDALTK-----CENDVNVIHAVARLFWRDKKIAKARKWMNRSVTLDPSLGD 211
N A L+K + ++ ARL +I +A+ + R T PS+
Sbjct 582 SANGNAEIARLILSKGYSSSLNDKEEIVLEAARLELSQGEIERAKIILERERTNSPSV-Q 640
Query 212 AWASYLAFELENGGEKECI 230
W + E + CI
Sbjct 641 IWVESIKLENDQKNYDLCI 659
Score = 40.8 bits (94), Expect = 0.006, Method: Composition-based stats.
Identities = 26/87 (29%), Positives = 40/87 (45%), Gaps = 1/87 (1%)
Query 80 LWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAI 139
W+ W N E N+ + VL+K+ P+ LWL Q + +GN E+A+ + +K
Sbjct 539 FWIKWANFEEKYGNFEKVDHVLQKSLKNCPDKQILWLKAAQNQSANGNAEIARLILSKGY 598
Query 140 QACPN-AGIIWAEAIFLEEENAQTHRA 165
+ N I EA LE + RA
Sbjct 599 SSSLNDKEEIVLEAARLELSQGEIERA 625
Score = 39.3 bits (90), Expect = 0.014, Method: Composition-based stats.
Identities = 33/129 (25%), Positives = 58/129 (44%), Gaps = 20/129 (15%)
Query 75 PRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHV 134
P S+ LW ++ ++ +A+L KA VP S ELWL ++ AQ +
Sbjct 364 PNSIKLWKEAISLVDNESE----KALLSKAVKCVPQSEELWLRYARLSEYCD----AQKI 415
Query 135 AAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCENDVNVIHAVARLFWRDKKIAK 194
+A + P IW EA LEE+N + + + +C ++++ K+
Sbjct 416 LNEARKVLPTFPGIWVEAAKLEEQNGKVEKVELIVKRCISNLSA-----------KRFVH 464
Query 195 AR-KWMNRS 202
+R W+NR+
Sbjct 465 SRDDWLNRA 473
> pfa:PF11_0108 U5 snRNP-associated protein, putative; K12855
pre-mRNA-processing factor 6
Length=1329
Score = 150 bits (378), Expect = 6e-36, Method: Composition-based stats.
Identities = 89/268 (33%), Positives = 146/268 (54%), Gaps = 16/268 (5%)
Query 5 NTANVWMQSIQLERQLGDYDAAIALADEA---VKQHQTF------AKLWMVRGQLHEEHP 55
++ N+W+ +I L+ + +Y A AL ++A +K +F K + + +
Sbjct 951 SSINLWICAIDLQIEKKNYTGARALTEKAKIKIKYLNSFNNNSHILKSKEIIETNEQNYD 1010
Query 56 TQRDLARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRV-PNSPEL 114
TQ D ++ +G+ + N +S+ + +V A +++ N L
Sbjct 1011 TQDDEYNNLNKNMDGSKSVNNTTA-----SNISKSKNELEKKSSVNNNAYIKIIENYDLL 1065
Query 115 WLALVQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCEN 174
WL L++IE+ N+ + + ++A++ CP++GI+W++AI LE +N Q ++V A C N
Sbjct 1066 WLKLIEIELCCNNKNL-NPIISEALKECPSSGILWSKAIELENKNLQNSKSVSAFNHCGN 1124
Query 175 DVNVIHAVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELENGGEKECIDIIN 234
+ VI VA+LFW + KI KARKW R + L+P GD WA++LAFE++ E DIIN
Sbjct 1125 NAYVILTVAKLFWVNFKIQKARKWFYRVINLNPHFGDGWATFLAFEIDQQNEINQKDIIN 1184
Query 235 RCALAQPNRGLAWNKTTKQVRCWSLSLP 262
+C A+PNRG WNK TK+V W L P
Sbjct 1185 KCIKAEPNRGYLWNKITKRVENWRLKYP 1212
Score = 63.9 bits (154), Expect = 5e-10, Method: Composition-based stats.
Identities = 60/292 (20%), Positives = 120/292 (41%), Gaps = 63/292 (21%)
Query 8 NVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAIDVF 67
++W+++++LE +L + +A +A+K T KLW+ E + ++++ V
Sbjct 459 DIWLEAVRLEEKLSEVKIILA---KAIKHIPTSVKLWL------EAYKKEKNVDDKRKVL 509
Query 68 REGTVCCPRSVPLWLCWVNCERSQK--------------------------NWNRARAVL 101
R+ C P SV LW ++ E + A+ VL
Sbjct 510 RKAIECIPNSVKLWKEAISLENENNAYILLKRAVECIPQSIEMWIALARLCTYTEAQKVL 569
Query 102 EKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQACPNAGIIW--AEAIFLEEEN 159
+A+ ++P S E+W+ Q+E + GN ++ + + I+ + +I+ + I EE
Sbjct 570 NEARKKIPTSAEIWINASQLEEKQGNIKMVDIIIKRCIENLSSKNVIFDRDKWIKFAEEC 629
Query 160 AQT--------------HRAVDALTK----CENDVNVIHAVARLFWRDKKIAKARKWMNR 201
Q+ H V+ L K ++ N IH +K I AR N
Sbjct 630 EQSKFTHTCESIIRNTMHIGVETLNKKRIYKQDAQNCIH--------NKSIHTARTLYNE 681
Query 202 SVTLDPSLGDAWASYLAFELENGGEKECIDIINRCALAQPNRGLAWNKTTKQ 253
++ + + W + EL +G ++ ++++R + P+ + W KQ
Sbjct 682 ALKIFKTKKSLWLALANLELTHGKREDVDEVLHRAVQSCPHSSVLWLMLAKQ 733
Score = 55.8 bits (133), Expect = 2e-07, Method: Composition-based stats.
Identities = 27/53 (50%), Positives = 35/53 (66%), Gaps = 0/53 (0%)
Query 1 RQNINTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEE 53
R NT +WMQS+QLER L +Y A LA EA+K H+ F KL+M+ GQ+ E
Sbjct 785 RVQCNTPKIWMQSVQLERLLRNYKEAKMLAHEALKIHKHFDKLYMIAGQIELE 837
Score = 50.4 bits (119), Expect = 7e-06, Method: Composition-based stats.
Identities = 38/155 (24%), Positives = 69/155 (44%), Gaps = 4/155 (2%)
Query 2 QNINTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLA 61
+ +N ++ Q Q A L +EA+K +T LW+ L H + D+
Sbjct 651 ETLNKKRIYKQDAQNCIHNKSIHTARTLYNEALKIFKTKKSLWLALANLELTHGKREDVD 710
Query 62 RAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQI 121
+V CP S LWL + ++AR +L ++ + N+ E+ LA +++
Sbjct 711 ---EVLHRAVQSCPHSSVLWLMLAKQKWLNNEIDKAREILAESFIHNQNTEEISLAAIKL 767
Query 122 EIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLE 156
E E+ + A+ + K+ C N IW +++ LE
Sbjct 768 ERENNEFDRARFLLKKSRVQC-NTPKIWMQSVQLE 801
Score = 43.9 bits (102), Expect = 6e-04, Method: Composition-based stats.
Identities = 47/209 (22%), Positives = 86/209 (41%), Gaps = 38/209 (18%)
Query 59 DLARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLAL 118
D+ +A + + P+ P W+ E + ++A+ ++ K + + ++WL
Sbjct 405 DINKARSLLKSVISTNPKHGPGWIAAARIEELAQRKDKAKEIIMKGCVVCSKNEDIWLEA 464
Query 119 VQIE------------------------IESGNREV----AQHVAAKAIQACPNAGIIWA 150
V++E +E+ +E + V KAI+ PN+ +W
Sbjct 465 VRLEEKLSEVKIILAKAIKHIPTSVKLWLEAYKKEKNVDDKRKVLRKAIECIPNSVKLWK 524
Query 151 EAIFLEEENAQ---THRAVDALTKCENDVNVIHAVARLFWRDKKIAKARKWMNRSVTLDP 207
EAI LE EN RAV+ + + + + A+ARL +A+K +N + P
Sbjct 525 EAISLENENNAYILLKRAVECIPQ---SIEMWIALARLC----TYTEAQKVLNEARKKIP 577
Query 208 SLGDAWASYLAFELENGGEKECIDIINRC 236
+ + W + E + G K II RC
Sbjct 578 TSAEIWINASQLEEKQGNIKMVDIIIKRC 606
Score = 42.7 bits (99), Expect = 0.001, Method: Composition-based stats.
Identities = 17/46 (36%), Positives = 31/46 (67%), Gaps = 0/46 (0%)
Query 63 AIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRV 108
A +++ EG C S+ LW+C ++ + +KN+ ARA+ EKAK+++
Sbjct 938 AQNIYEEGLKYCASSINLWICAIDLQIEKKNYTGARALTEKAKIKI 983
Score = 35.4 bits (80), Expect = 0.21, Method: Composition-based stats.
Identities = 29/98 (29%), Positives = 41/98 (41%), Gaps = 0/98 (0%)
Query 80 LWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAI 139
LWL N E + VL +A P+S LWL L + + + + A+ + A++
Sbjct 692 LWLALANLELTHGKREDVDEVLHRAVQSCPHSSVLWLMLAKQKWLNNEIDKAREILAESF 751
Query 140 QACPNAGIIWAEAIFLEEENAQTHRAVDALTKCENDVN 177
N I AI LE EN + RA L K N
Sbjct 752 IHNQNTEEISLAAIKLERENNEFDRARFLLKKSRVQCN 789
> bbo:BBOV_IV009720 23.m06014; u5 snRNP-associated subunit, putaitve;
K12855 pre-mRNA-processing factor 6
Length=1040
Score = 142 bits (357), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 68/149 (45%), Positives = 95/149 (63%), Gaps = 0/149 (0%)
Query 114 LWLALVQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCE 173
LWL V IE+ESGN A + + ++Q P++G +WA AIFLEE NAQ +AVDAL +C
Sbjct 850 LWLRAVDIELESGNAGNAYFMMSSSLQEFPDSGNLWARAIFLEERNAQNSKAVDALNQCS 909
Query 174 NDVNVIHAVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELENGGEKECIDII 233
N V+ A A+LFWRD K+ K RKW R++ ++ S G W ++LAFEL++G D+I
Sbjct 910 NSPLVVMAAAKLFWRDGKVLKTRKWFKRALAIEESNGVIWGTFLAFELDSGDNDAIKDVI 969
Query 234 NRCALAQPNRGLAWNKTTKQVRCWSLSLP 262
N C A+P+ G W + K+V W L+ P
Sbjct 970 NGCTKAEPSTGYDWCRVVKRVVNWRLTWP 998
Score = 95.5 bits (236), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 76/240 (31%), Positives = 114/240 (47%), Gaps = 11/240 (4%)
Query 1 RQNINTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDL 60
R+ T +WMQSIQLERQLG+Y AI L D+A++ H F KLWM+ GQL E P + D+
Sbjct 678 RKQCGTRKIWMQSIQLERQLGNYSVAIDLCDQALEIHPYFDKLWMIAGQLRLELP-EPDV 736
Query 61 ARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLE--KAKLRVPNSPELWLAL 118
A AI++F++G CP SV LWL + +ARA+++ K K+R P L
Sbjct 737 ATAINIFKDGADQCPWSVGLWLLALESLVRDNEHAKARALVDAAKTKIRCILGPRLKHTE 796
Query 119 VQIEIESGNREVAQHV-AAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCENDVN 177
+ + + + A+A P A + EEE + L C D+
Sbjct 797 QVATVNTKKLSPTELLRLARAYGDIPAATTTSMQ----EEEELIESLCQNILKSC--DLL 850
Query 178 VIHAVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELENGGEKECIDIINRCA 237
+ AV + A M+ S+ P G+ WA + E N + +D +N+C+
Sbjct 851 WLRAV-DIELESGNAGNAYFMMSSSLQEFPDSGNLWARAIFLEERNAQNSKAVDALNQCS 909
Score = 50.8 bits (120), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 52/256 (20%), Positives = 109/256 (42%), Gaps = 30/256 (11%)
Query 6 TANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAID 65
+ +W+++ + E + D + A E + +LW L +E L RA++
Sbjct 381 SVKIWVEAARRESNVNDKRRILRKALEFIPNS---VRLWKDAISLEDETDAYVMLKRAVE 437
Query 66 VFREGTVCCPRSVPLWLCWVN-CERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIE 124
C P SV LWL C ++ A+ VL +A+ +P + ++W+ ++E
Sbjct 438 -------CVPDSVDLWLALARLC-----SYQEAQKVLNEARKHLPTNADIWITAAKLEES 485
Query 125 SGNREVAQHVAAKAIQACPNAGIIWAEAIFLE-----EENAQTHRAVDALTKCENDVNVI 179
+GN+++ + + ++ + G+I + +L+ EEN A A+ KC ++ +
Sbjct 486 NGNQQMVEKIISRGLDNLSKKGVIHVRSNWLKQAEQCEENNFVQTA-QAIIKCTMNIGLD 544
Query 180 HAVARLFWRD--------KKIAKARKWMNRSVTLDPSLGDAWASYLAFELENGGEKECID 231
A+ + W + K A AR ++ + W + E +G ++ D
Sbjct 545 PALLKETWLEDGERMEEKKLFACARAIYRSALEQMKTKKSLWLALAELETRHGKPEDVDD 604
Query 232 IINRCALAQPNRGLAW 247
++++ PN + W
Sbjct 605 VLSQATKYCPNSDILW 620
Score = 48.1 bits (113), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 41/159 (25%), Positives = 71/159 (44%), Gaps = 4/159 (2%)
Query 9 VWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAIDVFR 68
W++ + + + A A+ A++Q +T LW+ +L H D+ DV
Sbjct 551 TWLEDGERMEEKKLFACARAIYRSALEQMKTKKSLWLALAELETRHGKPEDVD---DVLS 607
Query 69 EGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNR 128
+ T CP S LWL + Q + ARA+L A + + + LA V++E E
Sbjct 608 QATKYCPNSDILWLMAAKHKWIQGDVESARAILADAYSKNMDVESISLAAVKLEREHDEF 667
Query 129 EVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVD 167
E A+ + ++ + C IW ++I LE + A+D
Sbjct 668 ERARALLERSRKQCGTRK-IWMQSIQLERQLGNYSVAID 705
Score = 43.5 bits (101), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 31/110 (28%), Positives = 55/110 (50%), Gaps = 4/110 (3%)
Query 97 ARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLE 156
ARA+ A ++ LWLAL ++E G E V ++A + CPN+ I+W A +
Sbjct 568 ARAIYRSALEQMKTKKSLWLALAELETRHGKPEDVDDVLSQATKYCPNSDILWLMAAKHK 627
Query 157 --EENAQTHRAV--DALTKCENDVNVIHAVARLFWRDKKIAKARKWMNRS 202
+ + ++ RA+ DA +K + ++ A +L + +AR + RS
Sbjct 628 WIQGDVESARAILADAYSKNMDVESISLAAVKLEREHDEFERARALLERS 677
Score = 42.4 bits (98), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 36/147 (24%), Positives = 68/147 (46%), Gaps = 14/147 (9%)
Query 3 NINTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLAR 62
N N A W+ + ++E G +A + +A ++ +W+ +L + + LA+
Sbjct 314 NPNHAPGWIAAARIEELAGKISSAREIIAQACEKCGDREDVWLEAARLEKPEYAKAVLAK 373
Query 63 AIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIE 122
A+ + P+SV + WV R + N N R +L KA +PNS LW + +E
Sbjct 374 AVRM-------VPQSVKI---WVEAARRESNVNDKRRILRKALEFIPNSVRLWKDAISLE 423
Query 123 IESGNREVAQHVAAKAIQACPNAGIIW 149
E+ A + +A++ P++ +W
Sbjct 424 DETD----AYVMLKRAVECVPDSVDLW 446
Score = 38.9 bits (89), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 43/204 (21%), Positives = 82/204 (40%), Gaps = 31/204 (15%)
Query 59 DLARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLAL 118
D+ +A + + P P W+ E + AR ++ +A + + ++WL
Sbjct 299 DIKKARKLLKSVIATNPNHAPGWIAAARIEELAGKISSAREIIAQACEKCGDREDVWLEA 358
Query 119 VQIE-----------------------IESGNREV----AQHVAAKAIQACPNAGIIWAE 151
++E +E+ RE + + KA++ PN+ +W +
Sbjct 359 ARLEKPEYAKAVLAKAVRMVPQSVKIWVEAARRESNVNDKRRILRKALEFIPNSVRLWKD 418
Query 152 AIFLEEENAQTHRAVDALTKCENDVNVIHAVARLFWRDKKIAKARKWMNRSVTLDPSLGD 211
AI LE+E A+ + V++ A+ARL +A+K +N + P+ D
Sbjct 419 AISLEDETDAYVMLKRAVECVPDSVDLWLALARLC----SYQEAQKVLNEARKHLPTNAD 474
Query 212 AWASYLAFELENGGEKECIDIINR 235
W + E NG ++ II+R
Sbjct 475 IWITAAKLEESNGNQQMVEKIISR 498
Score = 35.4 bits (80), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 36/166 (21%), Positives = 67/166 (40%), Gaps = 5/166 (3%)
Query 60 LARAIDVFREGTVCCPRSVPLWLCWVNCERSQK--NWNRARAVLEKAKLRVPNSPELWLA 117
L + +D TV P+ L +N + + +AR +L+ PN W+A
Sbjct 264 LDKVMDNISGQTVVDPKGYLTDLNSMNIKSDSDIADIKKARKLLKSVIATNPNHAPGWIA 323
Query 118 LVQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCENDVN 177
+IE +G A+ + A+A + C + +W EA LE+ A+ V
Sbjct 324 AARIEELAGKISSAREIIAQACEKCGDREDVWLEAARLEKPEYAKAVLAKAVRMVPQSVK 383
Query 178 VIHAVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELEN 223
+ AR R+ + R+ + +++ P+ W ++ E E
Sbjct 384 IWVEAAR---RESNVNDKRRILRKALEFIPNSVRLWKDAISLEDET 426
Score = 33.9 bits (76), Expect = 0.68, Method: Compositional matrix adjust.
Identities = 38/169 (22%), Positives = 79/169 (46%), Gaps = 9/169 (5%)
Query 5 NTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAI 64
N+ +W+ + + + GD ++A A+ +A ++ + + +L EH + + ARA+
Sbjct 615 NSDILWLMAAKHKWIQGDVESARAILADAYSKNMDVESISLAAVKLEREH-DEFERARAL 673
Query 65 DVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIE 124
+ R C R + W+ + ER N++ A + ++A P +LW+ Q+ +E
Sbjct 674 -LERSRKQCGTRKI--WMQSIQLERQLGNYSVAIDLCDQALEIHPYFDKLWMIAGQLRLE 730
Query 125 SGNREVAQ--HVAAKAIQACPNAGIIW---AEAIFLEEENAQTHRAVDA 168
+VA ++ CP + +W E++ + E+A+ VDA
Sbjct 731 LPEPDVATAINIFKDGADQCPWSVGLWLLALESLVRDNEHAKARALVDA 779
Score = 30.4 bits (67), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 32/153 (20%), Positives = 67/153 (43%), Gaps = 23/153 (15%)
Query 9 VWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAIDVFR 68
+W++++ +E + G+ A + ++++ LW R EE Q ++A+D
Sbjct 850 LWLRAVDIELESGNAGNAYFMMSSSLQEFPDSGNLW-ARAIFLEERNAQN--SKAVDALN 906
Query 69 EGTVCCPRSVPLW------LCWVNCE--RSQKNWNRARAVLEKAKLRVPNSPELWLALVQ 120
+ C S PL L W + + +++K + RA A+ E + +W +
Sbjct 907 Q----CSNS-PLVVMAAAKLFWRDGKVLKTRKWFKRALAIEESNGV-------IWGTFLA 954
Query 121 IEIESGNREVAQHVAAKAIQACPNAGIIWAEAI 153
E++SG+ + + V +A P+ G W +
Sbjct 955 FELDSGDNDAIKDVINGCTKAEPSTGYDWCRVV 987
> sce:YBR055C PRP6, RNA6, TSM7269; Prp6p; K12855 pre-mRNA-processing
factor 6
Length=899
Score = 77.4 bits (189), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 53/219 (24%), Positives = 102/219 (46%), Gaps = 10/219 (4%)
Query 42 KLWMVRGQLHEEHPTQRDLARAIDVFREGTVCCPRSVPLWLCWVNC-ERSQKNWNRARAV 100
K ++ GQ++ + ++ + + + GT P LW+ E KN RAR++
Sbjct 669 KFFLQLGQIYH---SMGNIEMSRETYLSGTRLVPNCPLLWVSLSKIDEIDLKNPVRARSI 725
Query 101 LEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAI--FLEEE 158
L++ L+ P+ ++A +Q+EI GN + A+ + +A+Q P+ ++W E I F
Sbjct 726 LDRGLLKNPDDVLFYIAKIQMEIRLGNLDQAELLVTQALQKFPSNALLWVEQIKLFKHGN 785
Query 159 NAQTHRAV--DALTKCENDVNVIHAVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASY 216
+ + + DAL + +ND V+ + F+ + + + KW+ R++ GD W
Sbjct 786 KSSLKKTIFQDALRRTQNDHRVLLEIGVSFYAEAQYETSLKWLERALKKCSRYGDTWVWL 845
Query 217 LAFELENGGEKECIDIINRCALAQPNRGLAWNKTTKQVR 255
G K+ +D+ N +P G W +K V+
Sbjct 846 FRTYARLG--KDTVDLYNMFDQCEPTYGPEWIAASKNVK 882
Score = 38.9 bits (89), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 36/162 (22%), Positives = 66/162 (40%), Gaps = 6/162 (3%)
Query 56 TQRDLARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELW 115
T DL + + + P + W+ E + ++ A+ ++E P S ++W
Sbjct 219 TLEDLQKMRTILKSYRKADPTNPQGWIASARLEEKARKFSVAKKIIENGCQECPRSSDIW 278
Query 116 LALVQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAV-DALTKCEN 174
L I + + + + A AI P + ++W +AI LE +R V AL +
Sbjct 279 LE--NIRLHESDVHYCKTLVATAINFNPTSPLLWFKAIDLESTTVNKYRVVRKALQEIPR 336
Query 175 DVNVIHAVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASY 216
D + +A F DK A+ K + ++ P D +Y
Sbjct 337 DEG-LWKLAVSFEADK--AQVIKMLEKATQFIPQSMDLLTAY 375
Score = 30.4 bits (67), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 15/64 (23%), Positives = 31/64 (48%), Gaps = 4/64 (6%)
Query 59 DLARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLAL 118
D A+ I + + T P+S+ L + N + +++ A+ L + +P PE+W+
Sbjct 350 DKAQVIKMLEKATQFIPQSMDLLTAYTNLQ----SYHNAKMTLNSFRKILPQEPEIWIIS 405
Query 119 VQIE 122
+E
Sbjct 406 TLLE 409
> ath:AT5G41770 crooked neck protein, putative / cell cycle protein,
putative; K12869 crooked neck
Length=705
Score = 49.7 bits (117), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 46/178 (25%), Positives = 81/178 (45%), Gaps = 16/178 (8%)
Query 1 RQNINTANVWMQSIQLERQLGD-------YDAAIALADEAVKQH--QTFAKLWMVRGQLH 51
R++ + + W ++LE +G+ Y+ AIA A ++ Q + LW + L
Sbjct 335 RKSPSNYDSWFDYVRLEESVGNKDRIREIYERAIANVPPAEEKRYWQRYIYLW-INYALF 393
Query 52 EEHPTQRDLARAIDVFREGTVCCPRS----VPLWLCWVNCERSQKNWNRARAVLEKAKLR 107
EE T+ D+ R DV+RE P S +WL E Q N AR +L A +
Sbjct 394 EEIETE-DIERTRDVYRECLKLIPHSKFSFAKIWLLAAQFEIRQLNLTGARQILGNAIGK 452
Query 108 VPNSPELWLALVQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRA 165
P +++ ++IE++ GN + + + + ++ P W++ LE +T RA
Sbjct 453 APKD-KIFKKYIEIELQLGNMDRCRKLYERYLEWSPENCYAWSKYAELERSLVETERA 509
Score = 40.0 bits (92), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 52/211 (24%), Positives = 86/211 (40%), Gaps = 11/211 (5%)
Query 42 KLWMVRGQLHEEHPTQRDLARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVL 101
++W+ Q E +Q+D ARA V+ R+ LWL + E K N AR V
Sbjct 93 QVWVKYAQWEE---SQKDYARARSVWERAIEGDYRNHTLWLKYAEFEMKNKFVNSARNVW 149
Query 102 EKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQACPNA----GIIWAEAIFLEE 157
++A +P +LW + +E GN A+ + + + P+ I E + E
Sbjct 150 DRAVTLLPRVDQLWYKYIHMEEILGNIAGARQIFERWMDWSPDQQGWLSFIKFELRYNEI 209
Query 158 ENAQTHRAVDALTKCENDVNVIHAVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYL 217
E A+T + C V+ A+ + ++A+ R R+ +A ++
Sbjct 210 ERART--IYERFVLCHPKVSAYIRYAKFEMKGGEVARCRSVYERATEKLADDEEAEILFV 267
Query 218 AF-ELENG-GEKECIDIINRCALAQPNRGLA 246
AF E E E E I + AL +G A
Sbjct 268 AFAEFEERCKEVERARFIYKFALDHIPKGRA 298
Score = 37.0 bits (84), Expect = 0.064, Method: Compositional matrix adjust.
Identities = 52/232 (22%), Positives = 99/232 (42%), Gaps = 13/232 (5%)
Query 8 NVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAIDVF 67
VW++ Q E DY A ++ + A++ LW+ + ++ + + A +V+
Sbjct 93 QVWVKYAQWEESQKDYARARSVWERAIEGDYRNHTLWLKYAEFEMKN---KFVNSARNVW 149
Query 68 REGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGN 127
PR LW +++ E N AR + E+ P+ + WL+ ++ E+
Sbjct 150 DRAVTLLPRVDQLWYKYIHMEEILGNIAGARQIFERWMDWSPDQ-QGWLSFIKFELRYNE 208
Query 128 REVAQHVAAKAIQACP--NAGIIWAEAIFLEEENAQ----THRAVDALTKCENDVNVIHA 181
E A+ + + + P +A I +A+ E A+ RA + L E + A
Sbjct 209 IERARTIYERFVLCHPKVSAYIRYAKFEMKGGEVARCRSVYERATEKLADDEEAEILFVA 268
Query 182 VARLFWRDKKIAKARKWMNRSVTLDPS--LGDAWASYLAFELENGGEKECID 231
A R K++ +AR ++ P D + ++AFE + G+KE I+
Sbjct 269 FAEFEERCKEVERARFIYKFALDHIPKGRAEDLYRKFVAFE-KQYGDKEGIE 319
Score = 30.8 bits (68), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 40/187 (21%), Positives = 69/187 (36%), Gaps = 39/187 (20%)
Query 74 CPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPE---------LWL--ALVQIE 122
P + W +V E S N +R R + E+A VP + E LW+ AL + E
Sbjct 337 SPSNYDSWFDYVRLEESVGNKDRIREIYERAIANVPPAEEKRYWQRYIYLWINYALFE-E 395
Query 123 IESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCENDVNVIHAV 182
IE+ + E + V + ++ P++ +A+ L
Sbjct 396 IETEDIERTRDVYRECLKLIPHSKFSFAKIWLL--------------------------A 429
Query 183 ARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELENGGEKECIDIINRCALAQPN 242
A+ R + AR+ + ++ P + Y+ EL+ G C + R P
Sbjct 430 AQFEIRQLNLTGARQILGNAIGKAPK-DKIFKKYIEIELQLGNMDRCRKLYERYLEWSPE 488
Query 243 RGLAWNK 249
AW+K
Sbjct 489 NCYAWSK 495
> tpv:TP02_0476 crooked neck protein; K12869 crooked neck
Length=657
Score = 46.6 bits (109), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 22/80 (27%), Positives = 43/80 (53%), Gaps = 0/80 (0%)
Query 78 VPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAK 137
+ W+ + E +Q+ + RAR++ E+A L PN+P LWL ++ E+++ N A+++ +
Sbjct 73 IGTWIKYAVWEANQQEFRRARSIFERALLVDPNNPSLWLRYIETEMKNKNINSARNLFDR 132
Query 138 AIQACPNAGIIWAEAIFLEE 157
+ P W + EE
Sbjct 133 VVCLLPRIDQFWFKYAHFEE 152
Score = 43.1 bits (100), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 38/160 (23%), Positives = 66/160 (41%), Gaps = 5/160 (3%)
Query 56 TQRDLARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELW 115
Q++ RA +F + P + LWL ++ E KN N AR + ++ +P + W
Sbjct 85 NQQEFRRARSIFERALLVDPNNPSLWLRYIETEMKNKNINSARNLFDRVVCLLPRIDQFW 144
Query 116 LALVQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTK-CEN 174
E GN A+ + + ++ P W I EE + R + EN
Sbjct 145 FKYAHFEELLGNYAGARSIYERWMEWNPEDK-AWMLYIKFEERCGEVDRCRSIFNRYIEN 203
Query 175 DVNVIHAVARLFWRD--KKIAKARKWMNRSV-TLDPSLGD 211
+ + + + + + KK ++AR + V LDP L D
Sbjct 204 RPSCMSFLKLVKFEEKYKKTSRARSAFVKCVEVLDPELLD 243
Score = 32.0 bits (71), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 25/121 (20%), Positives = 52/121 (42%), Gaps = 2/121 (1%)
Query 52 EEHPTQRDLARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNS 111
E +++ A ++F PR W + + E N+ AR++ E+ P
Sbjct 115 ETEMKNKNINSARNLFDRVVCLLPRIDQFWFKYAHFEELLGNYAGARSIYERWMEWNPED 174
Query 112 PELWLALVQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTK 171
+ W+ ++ E G + + + + I+ P+ + + + + EE+ +T RA A K
Sbjct 175 -KAWMLYIKFEERCGEVDRCRSIFNRYIENRPSC-MSFLKLVKFEEKYKKTSRARSAFVK 232
Query 172 C 172
C
Sbjct 233 C 233
> ath:AT5G45990 crooked neck protein, putative / cell cycle protein,
putative
Length=673
Score = 46.6 bits (109), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 36/153 (23%), Positives = 70/153 (45%), Gaps = 4/153 (2%)
Query 77 SVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAA 136
++ +W+ + E SQ ++ RAR+V E+A + LW+ + E+++ A++V
Sbjct 77 NIQVWVKYAKWEESQMDYARARSVWERALEGEYRNHTLWVKYAEFEMKNKFVNNARNVWD 136
Query 137 KAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCEN---DVNVIHAVARLFWRDKKIA 193
+++ P +W + I++EE+ A + N D + R +I
Sbjct 137 RSVTLLPRVDQLWEKYIYMEEKLGNVTGARQIFERWMNWSPDQKAWLCFIKFELRYNEIE 196
Query 194 KARKWMNRSVTLDPSLGDAWASYLAFELENGGE 226
+AR R V P + A+ Y FE++ GG+
Sbjct 197 RARSIYERFVLCHPKVS-AFIRYAKFEMKRGGQ 228
Score = 35.4 bits (80), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 47/186 (25%), Positives = 77/186 (41%), Gaps = 22/186 (11%)
Query 75 PRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPE---------LWL--ALVQIEI 123
P + W +V E S N +R R + E+A VP + E LW+ AL + EI
Sbjct 325 PLNYDSWFDYVRLEESVGNKDRIREIYERAIANVPPAQEKRFWQRYIYLWINYALYE-EI 383
Query 124 ESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEE------NAQTHRAV--DALTKCEND 175
E+ + E + V + ++ P+ +A+ L E N R + +A+ K
Sbjct 384 ETKDVERTRDVYRECLKLIPHTKFSFAKIWLLAAEYEIRQLNLTGARQILGNAIGKAPK- 442
Query 176 VNVIHAVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELENGGEKECIDIINR 235
V + + + I + RK R + P AW +Y FE+ + E E I
Sbjct 443 VKIFKKYIEMELKLVNIDRCRKLYERFLEWSPENCYAWRNYAEFEI-SLAETERARAIFE 501
Query 236 CALAQP 241
A++QP
Sbjct 502 LAISQP 507
> hsa:51340 CRNKL1, CLF, CRN, Clf1, HCRN, MSTP021, SYF3; crooked
neck pre-mRNA splicing factor-like 1 (Drosophila); K12869
crooked neck
Length=848
Score = 45.4 bits (106), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 55/237 (23%), Positives = 94/237 (39%), Gaps = 18/237 (7%)
Query 12 QSIQLERQLGDYDA-AIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAIDVFREG 70
Q I E +L DY ++ +++++T W+ Q E + +++ RA ++
Sbjct 212 QKITDEEELNDYKLRKRKTFEDNIRKNRTVISNWIKYAQWEE---SLKEIQRARSIYERA 268
Query 71 TVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNREV 130
R++ LWL + E + N AR + ++A +P + W +E GN
Sbjct 269 LDVDYRNITLWLKYAEMEMKNRQVNHARNIWDRAITTLPRVNQFWYKYTYMEEMLGNVAG 328
Query 131 AQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRA---VDALTKCENDVNVIHAVARLFW 187
A+ V + ++ P W I E + RA + DV AR
Sbjct 329 ARQVFERWMEWQPEEQ-AWHSYINFELRYKEVDRARTIYERFVLVHPDVKNWIKYARFEE 387
Query 188 RDKKIAKARKWMNRSVTLDPSLGDA------WASYLAFELENGGEKECIDIINRCAL 238
+ A ARK R+V GD + ++ FE EN E E + +I + AL
Sbjct 388 KHAYFAHARKVYERAVEF---FGDEHMDEHLYVAFAKFE-ENQKEFERVRVIYKYAL 440
> cpv:cgd7_3690 crooked neck protein HAT repeats ; K12869 crooked
neck
Length=736
Score = 45.4 bits (106), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 25/101 (24%), Positives = 46/101 (45%), Gaps = 0/101 (0%)
Query 57 QRDLARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWL 116
Q ++ + +F G + +V +W ++ E + N N AR + E+ +P E W+
Sbjct 115 QNNIKNSRSIFERGILVNYENVRIWREYIKLEITNGNINNARNLFERVTHLLPRIDEFWI 174
Query 117 ALVQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEE 157
+Q+E+ N +H+ K I P+ I + F EE
Sbjct 175 KYIQMELILKNYINVRHIYRKWIDWKPDPSIYIQYSKFEEE 215
Score = 36.2 bits (82), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 34/160 (21%), Positives = 70/160 (43%), Gaps = 12/160 (7%)
Query 56 TQRDLARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELW 115
T ++ A ++F T PR W+ ++ E KN+ R + K P+ P ++
Sbjct 148 TNGNINNARNLFERVTHLLPRIDEFWIKYIQMELILKNYINVRHIYRKWIDWKPD-PSIY 206
Query 116 LALVQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQ---THRAVDALTKC 172
+ + E E G + A+ V I + P+ + E I E+ + + + ++ L++
Sbjct 207 IQYSKFEEECGEIKSARGVMKDLIISYPDES-NFIEYIKFEQRHKNLFSSEQIINILSET 265
Query 173 ENDVN-------VIHAVARLFWRDKKIAKARKWMNRSVTL 205
D+N +++ +F +KKI +A K N + +
Sbjct 266 LIDINGSKITNLFFSSISDIFVEEKKIEEAIKLCNEGIKI 305
Score = 36.2 bits (82), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 35/167 (20%), Positives = 65/167 (38%), Gaps = 31/167 (18%)
Query 78 VPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAK 137
+ L+L + E Q N +R++ E+ L + +W +++EI +GN A+++ +
Sbjct 102 ISLYLSYAKWESLQNNIKNSRSIFERGILVNYENVRIWREYIKLEITNGNINNARNLFER 161
Query 138 AIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCENDVNVIHAVARLFWRDKKIAKARK 197
P W + I +E +N +NV H RK
Sbjct 162 VTHLLPRIDEFWIKYIQME-------------LILKNYINVRHI-------------YRK 195
Query 198 WMNRSVTLDPSLGDAWASYLAFELENGGEKECIDIINRCALAQPNRG 244
W++ DPS+ + Y FE E G K ++ ++ P+
Sbjct 196 WIDWKP--DPSI---YIQYSKFEEECGEIKSARGVMKDLIISYPDES 237
> hsa:22984 PDCD11, ALG-4, ALG4, KIAA0185, NFBP, RRP5; programmed
cell death 11; K14792 rRNA biogenesis protein RRP5
Length=1871
Score = 45.1 bits (105), Expect = 2e-04, Method: Composition-based stats.
Identities = 54/263 (20%), Positives = 100/263 (38%), Gaps = 58/263 (22%)
Query 30 ADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLAR-------------AIDVFREGTVCCPR 76
++E K HQ K +L E+ +++L+R + D F + P
Sbjct 1559 SEEDEKPHQATIKKSKKEREL-EKQKAEKELSRIEEALMDPGRQPESADDFDRLVLSSPN 1617
Query 77 SVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPE-----LWLALVQIEIESGNREVA 131
S LWL ++ +ARAV E+A + E +W+AL+ +E G++E
Sbjct 1618 SSILWLQYMAFHLQATEIEKARAVAERALKTISFREEQEKLNVWVALLNLENMYGSQESL 1677
Query 132 QHVAAKAIQ-------------------ACPNAG-----------------IIWAEAIFL 155
V +A+Q AG I + +
Sbjct 1678 TKVFERAVQYNEPLKVFLHLADIYAKSEKFQEAGELYNRMLKRFRQEKAVWIKYGAFLLR 1737
Query 156 EEENAQTHRAVDALTKC---ENDVNVIHAVARLFWRDKKIAKARKWMNRSVTLDPSLGDA 212
+ A +HR + +C + V+VI A+L ++ +A+ +++ P D
Sbjct 1738 RSQAAASHRVLQRALECLPSKEHVDVIAKFAQLEFQLGDAERAKAIFENTLSTYPKRTDV 1797
Query 213 WASYLAFELENGGEKECIDIINR 235
W+ Y+ +++G +K+ DI R
Sbjct 1798 WSVYIDMTIKHGSQKDVRDIFER 1820
> mmu:66877 Crnkl1, 1200013P10Rik, 5730590A01Rik, C80326, crn;
Crn, crooked neck-like 1 (Drosophila); K12869 crooked neck
Length=690
Score = 44.7 bits (104), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 59/240 (24%), Positives = 97/240 (40%), Gaps = 24/240 (10%)
Query 12 QSIQLERQLGDYDA-AIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAIDVFREG 70
Q I E +L DY ++ +++++T W+ Q E + +++ RA ++
Sbjct 51 QKITDEEELNDYKLRKRKTFEDNIRKNRTVISNWIKYAQWEE---SLKEIQRARSIYERA 107
Query 71 TVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNREV 130
R++ LWL + E + N AR + ++A +P + W +E GN
Sbjct 108 LDVDYRNITLWLKYAEMEMKNRQVNHARNIWDRAITTLPRVNQFWYKYTYMEEMLGNVAG 167
Query 131 AQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCENDVNVIHAV------AR 184
A+ V + ++ P W I E + R A T E V V AV AR
Sbjct 168 ARQVFERWMEWQPEEQ-AWHSYINFELRYKEVER---ARTIYERFVLVHPAVKNWIKYAR 223
Query 185 LFWRDKKIAKARKWMNRSVTLDPSLGDA------WASYLAFELENGGEKECIDIINRCAL 238
+ A ARK R+V GD + ++ FE EN E E + +I + AL
Sbjct 224 FEEKHAYFAHARKVYERAVEF---FGDEHMDEHLYVAFAKFE-ENQKEFERVRVIYKYAL 279
> bbo:BBOV_III004750 17.m07426; tetratricopeptide repeat domain
containing protein; K12869 crooked neck
Length=665
Score = 43.5 bits (101), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 21/77 (27%), Positives = 41/77 (53%), Gaps = 0/77 (0%)
Query 81 WLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQ 140
W+ + E +Q+++ RAR+V E+A PN+ LWL ++ E+++ N A+++ + +
Sbjct 76 WIKYALWEANQQDFRRARSVFERALQVDPNNVNLWLRYIETEMKNKNVNAARNLFDRVVS 135
Query 141 ACPNAGIIWAEAIFLEE 157
P W + EE
Sbjct 136 LLPRVDQFWFKYAHFEE 152
Score = 43.1 bits (100), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/88 (28%), Positives = 39/88 (44%), Gaps = 0/88 (0%)
Query 57 QRDLARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWL 116
Q+D RA VF P +V LWL ++ E KN N AR + ++ +P + W
Sbjct 86 QQDFRRARSVFERALQVDPNNVNLWLRYIETEMKNKNVNAARNLFDRVVSLLPRVDQFWF 145
Query 117 ALVQIEIESGNREVAQHVAAKAIQACPN 144
E GN A+ V + ++ P+
Sbjct 146 KYAHFEELLGNYAGARTVFERWMEWNPD 173
Score = 35.0 bits (79), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 37/149 (24%), Positives = 65/149 (43%), Gaps = 7/149 (4%)
Query 13 SIQLERQLGDYDAAIALADEAVK-QHQTFAKLWMVRGQLHEEHPTQRDLARAIDVFREGT 71
+I E L D A+A+ +A++ + FAK +++ +L+ Q DL F G
Sbjct 387 AIFSELTLQQLDRAVAVYRKALQVLPKDFAKFYILLAELY---LRQGDLDSMRKTFGLGL 443
Query 72 VCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNREVA 131
C + L+ + E N +R R + K P PE WL+ +++E+ R+
Sbjct 444 GQCKKP-KLFETYAQIELKLGNLDRCRHIHAKYIETWPFKPESWLSFIELELMLNERKRV 502
Query 132 QHV--AAKAIQACPNAGIIWAEAIFLEEE 158
+ + AA A+ +W I +E E
Sbjct 503 RGLCEAAIAMDQMDMPETVWNRYIEIERE 531
> cel:T20B12.1 hypothetical protein
Length=771
Score = 43.5 bits (101), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 40/152 (26%), Positives = 65/152 (42%), Gaps = 34/152 (22%)
Query 113 ELWLALVQIEIESGNREVAQHVAAKAIQACPNAGII-------------WAEAIFLEEE- 158
E+W ++ + G + A+ + + I+ PN ++ + +AI L ++
Sbjct 390 EMWDGVIDCYKQLGQMDKAETLIRRLIEQKPNDSMLHVYLGDITRNLEYFTKAIELSDDR 449
Query 159 NAQTHRAVDALTKCENDVNVIHAVARLFWRDKKIAKARKWMNRSVTLDPSLGDAW--ASY 216
NA+ HR++ L DKK +A K + RS+ L P W A Y
Sbjct 450 NARAHRSL----------------GHLLLMDKKFEEAYKHLRRSLELQPIQLGTWFNAGY 493
Query 217 LAFELENGGEKECIDIINRCALAQPNRGLAWN 248
A++LEN KE +RC QP+ AWN
Sbjct 494 CAWKLENF--KESTQCYHRCVSLQPDHFEAWN 523
> sce:YLR117C CLF1, NTC77, SYF3; Clf1p; K12869 crooked neck
Length=687
Score = 42.4 bits (98), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 21/81 (25%), Positives = 39/81 (48%), Gaps = 0/81 (0%)
Query 57 QRDLARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWL 116
Q D+ RA +F + +PLW+ +++ E K N AR ++ +A +P +LW
Sbjct 78 QHDMRRARSIFERALLVDSSFIPLWIRYIDAELKVKCINHARNLMNRAISTLPRVDKLWY 137
Query 117 ALVQIEIESGNREVAQHVAAK 137
+ +E N E+ + + K
Sbjct 138 KYLIVEESLNNVEIVRSLYTK 158
Score = 41.2 bits (95), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 39/205 (19%), Positives = 86/205 (41%), Gaps = 29/205 (14%)
Query 29 LADEAV-KQHQTFAKLWMVRGQLHEEHPTQRDLARAIDVFREGTVCCPRSVPLWLCWVNC 87
L D+ + +H TF+K+W++ + H D+ +A + + CP++ + ++
Sbjct 391 LIDDIIPHKHFTFSKIWLMYAKFLIRHD---DVPKARKILGKAIGLCPKA-KTFKGYIEL 446
Query 88 ERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQACPNAGI 147
E K ++R R + EK P+ ++W ++E G+ + + + A+
Sbjct 447 EVKLKEFDRVRKIYEKFIEFQPSDLQIWSQYGELEENLGDWDRVRGIYTIAL-------- 498
Query 148 IWAEAIFLEEENAQTHRAVDALTKCENDVNVIHAVARLFWRDKKIAKARKWMNRSVTLDP 207
+EN+ D LTK E + ++ ++ KARK R + L+
Sbjct 499 ---------DENS------DFLTK-EAKIVLLQKYITFETESQEFEKARKLYRRYLELNQ 542
Query 208 SLGDAWASYLAFELENGGEKECIDI 232
+W + ++ E++ +D+
Sbjct 543 YSPQSWIEFAMYQTSTPTEQQLLDL 567
Score = 39.3 bits (90), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 33/161 (20%), Positives = 68/161 (42%), Gaps = 31/161 (19%)
Query 81 WLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQ 140
W+ + E Q + RAR++ E+A L + LW+ + E++ A+++ +AI
Sbjct 68 WIRYAQFEIEQHDMRRARSIFERALLVDSSFIPLWIRYIDAELKVKCINHARNLMNRAIS 127
Query 141 ACPNAGIIWAEAIFLEEENAQTHRAVDALTKCENDVNVIHAVARLFWRDKKIAKARKWMN 200
P +W + + +EE N+V ++ ++ KW
Sbjct 128 TLPRVDKLWYKYLIVEE--------------SLNNVEIVRSL------------YTKW-- 159
Query 201 RSVTLDPSLGDAWASYLAFELENGGEKECIDIINRCALAQP 241
+L+P + +AW S++ FE+ +I ++ +A P
Sbjct 160 --CSLEPGV-NAWNSFVDFEIRQKNWNGVREIYSKYVMAHP 197
Score = 38.5 bits (88), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 31/68 (45%), Gaps = 1/68 (1%)
Query 77 SVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAA 136
V W +V+ E QKNWN R + K + P + WL V+ E GN E + V +
Sbjct 165 GVNAWNSFVDFEIRQKNWNGVREIYSKYVMAHPQM-QTWLKWVRFENRHGNTEFTRSVYS 223
Query 137 KAIQACPN 144
AI N
Sbjct 224 LAIDTVAN 231
Score = 30.0 bits (66), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 34/138 (24%), Positives = 63/138 (45%), Gaps = 18/138 (13%)
Query 9 VWMQSIQLERQLGDYDAAIALADEAVKQHQTF----AKLWMVRGQLHEEHPTQRDLARAI 64
+W Q +LE LGD+D + A+ ++ F AK+ +++ + E +Q + +A
Sbjct 473 IWSQYGELEENLGDWDRVRGIYTIALDENSDFLTKEAKIVLLQKYITFETESQ-EFEKAR 531
Query 65 DVFR---EGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQI 121
++R E P+S W+ Q + + +L+ AKL+ N E ++
Sbjct 532 KLYRRYLELNQYSPQS------WIEFAMYQTSTPTEQQLLDLAKLQSENVDE----DIEF 581
Query 122 EIESGNREVAQHVAAKAI 139
EI N+ A+ V +AI
Sbjct 582 EITDENKLEARKVFEEAI 599
> ath:AT3G17040 HCF107; HCF107 (HIGH CHLOROPHYLL FLUORESCENT 107);
binding
Length=652
Score = 41.6 bits (96), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 49/226 (21%), Positives = 89/226 (39%), Gaps = 9/226 (3%)
Query 16 LERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAIDVFREGTVCCP 75
LE +LG+ A L D A + W L + Q ++++A ++ +G C
Sbjct 246 LENRLGNVRRARELFDAATVADKKHVAAWHGWANLEIK---QGNISKARNLLAKGLKFCG 302
Query 76 RSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVA 135
R+ ++ E + +AR + ++A + S WLA Q+EI+ A+ +
Sbjct 303 RNEYIYQTLALLEAKAGRYEQARYLFKQATICNSRSCASWLAWAQLEIQQERYPAARKLF 362
Query 136 AKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCENDVN-----VIHAVARLFWRDK 190
KA+QA P W E R L K + +N ++ ++ L ++
Sbjct 363 EKAVQASPKNRFAWHVWGVFEAGVGNVERG-RKLLKIGHALNPRDPVLLQSLGLLEYKHS 421
Query 191 KIAKARKWMNRSVTLDPSLGDAWASYLAFELENGGEKECIDIINRC 236
AR + R+ LDP W ++ E + G ++ R
Sbjct 422 SANLARALLRRASELDPRHQPVWIAWGWMEWKEGNTTTARELYQRA 467
Score = 40.8 bits (94), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 34/146 (23%), Positives = 63/146 (43%), Gaps = 6/146 (4%)
Query 66 VFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIES 125
+F++ T+C RS WL W E Q+ + AR + EKA P + W E
Sbjct 327 LFKQATICNSRSCASWLAWAQLEIQQERYPAARKLFEKAVQASPKNRFAWHVWGVFEAGV 386
Query 126 GNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCENDVNVIH----- 180
GN E + + P ++ LE +++ + A AL + ++++ H
Sbjct 387 GNVERGRKLLKIGHALNPRDPVLLQSLGLLEYKHSSANLA-RALLRRASELDPRHQPVWI 445
Query 181 AVARLFWRDKKIAKARKWMNRSVTLD 206
A + W++ AR+ R++++D
Sbjct 446 AWGWMEWKEGNTTTARELYQRALSID 471
Score = 38.9 bits (89), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 36/146 (24%), Positives = 58/146 (39%), Gaps = 6/146 (4%)
Query 80 LWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAI 139
+W CW E N RAR + + A + W +EI+ GN A+++ AK +
Sbjct 239 IWQCWAVLENRLGNVRRARELFDAATVADKKHVAAWHGWANLEIKQGNISKARNLLAKGL 298
Query 140 QACPNAGIIWAEAIFLEEENAQTHRAVDALTKCENDVNVIHAVARLFW-----RDKKIAK 194
+ C I+ + + L E A + L K N + L W + ++
Sbjct 299 KFCGRNEYIY-QTLALLEAKAGRYEQARYLFKQATICNSRSCASWLAWAQLEIQQERYPA 357
Query 195 ARKWMNRSVTLDPSLGDAWASYLAFE 220
ARK ++V P AW + FE
Sbjct 358 ARKLFEKAVQASPKNRFAWHVWGVFE 383
Score = 33.5 bits (75), Expect = 0.77, Method: Compositional matrix adjust.
Identities = 32/122 (26%), Positives = 55/122 (45%), Gaps = 8/122 (6%)
Query 48 GQLHEEHPTQRDLARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLR 107
G L +H + +LARA+ R + PR P+W+ W E + N AR + ++A L
Sbjct 414 GLLEYKHSSA-NLARAL--LRRASELDPRHQPVWIAWGWMEWKEGNTTTARELYQRA-LS 469
Query 108 VPNSPELWLALVQ----IEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTH 163
+ + E +Q +E +GN A+ + ++ + + W LEE+ T
Sbjct 470 IDANTESASRCLQAWGVLEQRAGNLSAARRLFRSSLNINSQSYVTWMTWAQLEEDQGDTE 529
Query 164 RA 165
RA
Sbjct 530 RA 531
Score = 32.0 bits (71), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 35/136 (25%), Positives = 54/136 (39%), Gaps = 3/136 (2%)
Query 10 WMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAIDVFRE 69
W+ QLE Q Y AA L ++AV+ W V G ++ R + +
Sbjct 342 WLAWAQLEIQQERYPAARKLFEKAVQASPKNRFAWHVWGVF---EAGVGNVERGRKLLKI 398
Query 70 GTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNRE 129
G PR L E + N ARA+L +A P +W+A +E + GN
Sbjct 399 GHALNPRDPVLLQSLGLLEYKHSSANLARALLRRASELDPRHQPVWIAWGWMEWKEGNTT 458
Query 130 VAQHVAAKAIQACPNA 145
A+ + +A+ N
Sbjct 459 TARELYQRALSIDANT 474
> cel:M03F8.3 hypothetical protein; K12869 crooked neck
Length=747
Score = 41.6 bits (96), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 39/169 (23%), Positives = 71/169 (42%), Gaps = 9/169 (5%)
Query 39 TFAKLWMVRGQLHEEHPTQRDLARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRAR 98
TFAK+W++ Q DL A + CP+ L+ +++ E + ++R R
Sbjct 420 TFAKVWIMFAHFE---IRQLDLNAARKIMGVAIGKCPKD-KLFRAYIDLELQLREFDRCR 475
Query 99 AVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQ--ACPNAGIIWAEAIFLE 156
+ EK P S + W+ ++E G+ + ++ V A+Q A ++W I E
Sbjct 476 KLYEKFLESSPESSQTWIKFAELETLLGDTDRSRAVFTIAVQQPALDMPELLWKAYIDFE 535
Query 157 ---EENAQTHRAVDALTKCENDVNVIHAVARLFWRDKKIAKARKWMNRS 202
EE+ + + L + N + V ++A ARK R+
Sbjct 536 IACEEHEKARDLYETLLQRTNHIKVWISMAEFEQTIGNFEGARKAFERA 584
Score = 37.0 bits (84), Expect = 0.083, Method: Compositional matrix adjust.
Identities = 40/161 (24%), Positives = 63/161 (39%), Gaps = 3/161 (1%)
Query 67 FREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESG 126
F +G + W+ + E S RAR+V E+A S +WL ++E+
Sbjct 75 FEDGIRKNRMQLANWIKYGKWEESIGEIQRARSVFERALDVDHRSISIWLQYAEMEMRCK 134
Query 127 NREVAQHVAAKAIQACPNAGIIWAEAIFLEE--ENAQTHRAV-DALTKCENDVNVIHAVA 183
A++V +AI P A W + ++EE EN R + + + E
Sbjct 135 QINHARNVFDRAITIMPRAMQFWLKYSYMEEVIENIPGARQIFERWIEWEPPEQAWQTYI 194
Query 184 RLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELENG 224
R K+I +AR R + + W Y FE NG
Sbjct 195 NFELRYKEIDRARSVYQRFLHVHGINVQNWIKYAKFEERNG 235
Score = 35.8 bits (81), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 30/64 (46%), Gaps = 0/64 (0%)
Query 59 DLARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLAL 118
++ RA VF RS+ +WL + E K N AR V ++A +P + + WL
Sbjct 101 EIQRARSVFERALDVDHRSISIWLQYAEMEMRCKQINHARNVFDRAITIMPRAMQFWLKY 160
Query 119 VQIE 122
+E
Sbjct 161 SYME 164
Score = 32.0 bits (71), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 35/139 (25%), Positives = 59/139 (42%), Gaps = 17/139 (12%)
Query 6 TANVWMQSIQLERQLGDYDAAIALADEAVKQH--QTFAKLWMVRGQLH---EEHPTQRDL 60
++ W++ +LE LGD D + A+ AV+Q LW EEH RDL
Sbjct 488 SSQTWIKFAELETLLGDTDRSRAVFTIAVQQPALDMPELLWKAYIDFEIACEEHEKARDL 547
Query 61 ARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPE-----LW 115
E + + +W+ E++ N+ AR E+A + N+ + L
Sbjct 548 Y-------ETLLQRTNHIKVWISMAEFEQTIGNFEGARKAFERANQSLENAEKEERLMLL 600
Query 116 LALVQIEIESGNREVAQHV 134
A + E +SG++E + V
Sbjct 601 EAWKECETKSGDQEALKRV 619
> dre:794079 pdcd11, MGC162501, cb680, im:7148359, sb:cb680, wu:fc68c10;
programmed cell death 11; K14792 rRNA biogenesis
protein RRP5
Length=1816
Score = 41.2 bits (95), Expect = 0.004, Method: Composition-based stats.
Identities = 42/185 (22%), Positives = 79/185 (42%), Gaps = 46/185 (24%)
Query 96 RARAVLEKAKLRVPNSPE-----LWLALVQIEIESGNREVAQHVAAKAIQACP------N 144
+ARAV E+A + E +W+A++ +E G + Q V +AIQ C
Sbjct 1582 QARAVAERALKTISFREEQEKLNIWVAMLNLENMYGTPDSLQKVFERAIQYCEPLLVYQQ 1641
Query 145 AGIIWAE----------------------AIFL------------EEENAQTHRAVDALT 170
I+A+ A++L + NA RA+ +L+
Sbjct 1642 LADIYAKSEKIKEAESLYKSMVKRFRQDKAVYLSYGTFLLRQRQSDAANALLQRALQSLS 1701
Query 171 KCENDVNVIHAVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELENGGEKECI 230
E+ V++I ARL ++ KA+ ++ +T P D W+ ++ +++G +KE
Sbjct 1702 SKEH-VDLIARFARLEFQFGNSEKAKSMFDKVLTTYPKRTDLWSVFIDLMVKHGSQKEVR 1760
Query 231 DIINR 235
++ +R
Sbjct 1761 ELFDR 1765
Score = 32.7 bits (73), Expect = 1.5, Method: Composition-based stats.
Identities = 28/152 (18%), Positives = 64/152 (42%), Gaps = 6/152 (3%)
Query 1 RQNINTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDL 60
R+ N+W+ + LE G D+ + + A++ + + +V QL + + +
Sbjct 1597 REEQEKLNIWVAMLNLENMYGTPDSLQKVFERAIQ----YCEPLLVYQQLADIYAKSEKI 1652
Query 61 ARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKA--KLRVPNSPELWLAL 118
A +++ + ++L + Q+ + A A+L++A L +L
Sbjct 1653 KEAESLYKSMVKRFRQDKAVYLSYGTFLLRQRQSDAANALLQRALQSLSSKEHVDLIARF 1712
Query 119 VQIEIESGNREVAQHVAAKAIQACPNAGIIWA 150
++E + GN E A+ + K + P +W+
Sbjct 1713 ARLEFQFGNSEKAKSMFDKVLTTYPKRTDLWS 1744
> mmu:18572 Pdcd11, 1110021I22Rik, ALG-4, Pdcd7, mKIAA0185; programmed
cell death 11; K14792 rRNA biogenesis protein RRP5
Length=1862
Score = 40.4 bits (93), Expect = 0.007, Method: Composition-based stats.
Identities = 44/215 (20%), Positives = 83/215 (38%), Gaps = 44/215 (20%)
Query 65 DVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPE-----LWLALV 119
D F + P S LWL ++ +ARAV E+A + E +W+AL+
Sbjct 1597 DDFDRLVLSSPNSSILWLQYMAFHLQATEIEKARAVAERALKTISFREEQEKLNVWVALL 1656
Query 120 QIEIESGNREVAQHVAAKAIQ-------------------ACPNAGIIWAEAI--FLEE- 157
+E G++E V +A+Q AG ++ + F +E
Sbjct 1657 NLENMYGSQESLTKVFERAVQYNEPLKVFLHLADIYTKSEKYKEAGELYNRMLKRFRQEK 1716
Query 158 --------------ENAQTHRAVDALTKC---ENDVNVIHAVARLFWRDKKIAKARKWMN 200
+ +HR + +C + V+VI A+L ++ + +A+
Sbjct 1717 AVWIKYGAFVLGRSQAGASHRVLQRALECLPAKEHVDVIVKFAQLEFQLGDVERAKAIFE 1776
Query 201 RSVTLDPSLGDAWASYLAFELENGGEKECIDIINR 235
+++ P D W+ Y+ +++G + DI R
Sbjct 1777 NTLSTYPKRTDVWSVYIDMTIKHGSQTAVRDIFER 1811
> hsa:55015 PRPF39, FLJ11128, FLJ20666, FLJ45460, MGC149842, MGC149843;
PRP39 pre-mRNA processing factor 39 homolog (S. cerevisiae);
K13217 pre-mRNA-processing factor 39
Length=669
Score = 39.3 bits (90), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 32/132 (24%), Positives = 57/132 (43%), Gaps = 5/132 (3%)
Query 10 WMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAIDVF-R 68
W + ++ E + G ++ + L + V + + W+ + E H + VF R
Sbjct 356 WKEYLEFEIENGTHERVVVLFERCVISCALYEEFWIKYAKYMENH----SIEGVRHVFSR 411
Query 69 EGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNR 128
T+ P+ + + W E Q N N AR +L+ + V + L V +E GN
Sbjct 412 ACTIHLPKKPMVHMLWAAFEEQQGNINEARNILKTFEECVLGLAMVRLRRVSLERRHGNL 471
Query 129 EVAQHVAAKAIQ 140
E A+H+ AI+
Sbjct 472 EEAEHLLQDAIK 483
Score = 30.8 bits (68), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 10/27 (37%), Positives = 16/27 (59%), Gaps = 0/27 (0%)
Query 213 WASYLAFELENGGEKECIDIINRCALA 239
W YL FE+ENG + + + RC ++
Sbjct 356 WKEYLEFEIENGTHERVVVLFERCVIS 382
> sce:YBR112C CYC8, CRT8, SSN6; Cyc8p; K06665 glucose repression
mediator protein
Length=966
Score = 38.9 bits (89), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 34/143 (23%), Positives = 60/143 (41%), Gaps = 7/143 (4%)
Query 5 NTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDL-ARA 63
+TA W+ L LGD D A D ++ + + AK L+ RD+ RA
Sbjct 44 STAETWLSIASLAETLGDGDRAAMAYDATLQFNPSSAKALTSLAHLYRS----RDMFQRA 99
Query 64 IDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKA--KLRVPNSPELWLALVQI 121
+++ + P +W +C + RA ++A L PN P+LW + +
Sbjct 100 AELYERALLVNPELSDVWATLGHCYLMLDDLQRAYNAYQQALYHLSNPNVPKLWHGIGIL 159
Query 122 EIESGNREVAQHVAAKAIQACPN 144
G+ + A+ AK ++ P+
Sbjct 160 YDRYGSLDYAEEAFAKVLELDPH 182
Score = 30.8 bits (68), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 40/188 (21%), Positives = 73/188 (38%), Gaps = 3/188 (1%)
Query 8 NVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQ-RDLARAIDV 66
++W Q + +G++ A + + Q+Q AK+ G L+ Q D +A+D
Sbjct 225 DIWFQLGSVLESMGEWQGAKEAYEHVLAQNQHHAKVLQQLGCLYGMSNVQFYDPQKALDY 284
Query 67 FREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESG 126
+ P W + ++ A ++A R +P W ++ + +
Sbjct 285 LLKSLEADPSDATTWYHLGRVHMIRTDYTAAYDAFQQAVNRDSRNPIFWCSIGVLYYQIS 344
Query 127 NREVAQHVAAKAIQACPNAGIIWAEAIFLEEE-NAQTHRAVDALTKCEN-DVNVIHAVAR 184
A +AI+ P +W + L E N Q A+DA + DVN +H R
Sbjct 345 QYRDALDAYTRAIRLNPYISEVWYDLGTLYETCNNQLSDALDAYKQAARLDVNNVHIRER 404
Query 185 LFWRDKKI 192
L K++
Sbjct 405 LEALTKQL 412
> mmu:328110 Prpf39, FLJ11128, MGC37077, Srcs1; PRP39 pre-mRNA
processing factor 39 homolog (yeast); K13217 pre-mRNA-processing
factor 39
Length=665
Score = 38.5 bits (88), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 33/132 (25%), Positives = 55/132 (41%), Gaps = 5/132 (3%)
Query 10 WMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAIDVF-R 68
W + ++ E + G ++ + L + V + + W+ + E H + VF R
Sbjct 354 WKEYLEFEIENGTHERVVVLFERCVISCALYEEFWIKYAKYMENH----SIEGVRHVFSR 409
Query 69 EGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNR 128
TV P+ + W E Q N N AR +L + V + L V +E GN
Sbjct 410 ACTVHLPKKPMAHMLWAAFEEQQGNINEARIILRTFEECVLGLAMVRLRRVSLERRHGNM 469
Query 129 EVAQHVAAKAIQ 140
E A+H+ AI+
Sbjct 470 EEAEHLLQDAIK 481
Score = 31.6 bits (70), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 33/132 (25%), Positives = 51/132 (38%), Gaps = 13/132 (9%)
Query 10 WMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAIDVFRE 69
W+ +Q Q AA D+ + W L + H ++ ++ +V+R
Sbjct 96 WVYLLQYVEQENHLMAARKAFDKFFVHYPYCYGYWKKYADLEKRH---DNIKQSDEVYRR 152
Query 70 GTVCCPRSVPLWLCWVNCER------SQKNWNRARAVLEKAKLRVPN---SPELWLALVQ 120
G P SV LW+ ++N + Q+ R E A L S +LW +
Sbjct 153 GLQAIPLSVDLWIHYINFLKETLDPGDQETNTTIRGTFEHAVLAAGTDFRSDKLWEMYIN 212
Query 121 IEIESGN-REVA 131
E E GN REV
Sbjct 213 WENEQGNLREVT 224
Score = 30.8 bits (68), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 10/27 (37%), Positives = 16/27 (59%), Gaps = 0/27 (0%)
Query 213 WASYLAFELENGGEKECIDIINRCALA 239
W YL FE+ENG + + + RC ++
Sbjct 354 WKEYLEFEIENGTHERVVVLFERCVIS 380
> hsa:84899 TMTC4, FLJ14624, FLJ22153; transmembrane and tetratricopeptide
repeat containing 4
Length=741
Score = 37.7 bits (86), Expect = 0.039, Method: Compositional matrix adjust.
Identities = 34/141 (24%), Positives = 62/141 (43%), Gaps = 3/141 (2%)
Query 5 NTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAI 64
+ A WM ++ L ++AA A+K + + + G+L+ + D A+
Sbjct 548 DFAAAWMNLGIVQNSLKRFEAAEQSYRTAIKHRRKYPDCYYNLGRLYADLNRHVD---AL 604
Query 65 DVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIE 124
+ +R TV P W + + N +A AV +A +PN L +L + +
Sbjct 605 NAWRNATVLKPEHSLAWNNMIILLDNTGNLAQAEAVGREALELIPNDHSLMFSLANVLGK 664
Query 125 SGNREVAQHVAAKAIQACPNA 145
S + ++ + KAI+A PNA
Sbjct 665 SQKYKESEALFLKAIKANPNA 685
> xla:779090 pdcd11, alg-4, alg4, nfbp, rrp5; programmed cell
death 11; K14792 rRNA biogenesis protein RRP5
Length=1812
Score = 37.4 bits (85), Expect = 0.050, Method: Composition-based stats.
Identities = 46/219 (21%), Positives = 85/219 (38%), Gaps = 46/219 (21%)
Query 62 RAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPE-----LWL 116
++ D F + P S LWL ++ +AR V E+A + E +W+
Sbjct 1544 QSADDFDRLVISSPDSSILWLQYMAFHLHATEIEKARVVAERALKTISFREEQEKLNVWV 1603
Query 117 ALVQIEIESGNREVAQHVAAKAIQACPNAGI----------------------------- 147
AL+ +E G E +A+Q +
Sbjct 1604 ALLNLENMYGTEESLTKAFERAVQYNEPLKVFQQLADIYIKSEKFKQAEDLYNTMLKRFR 1663
Query 148 ----IWAE-AIFL--EEENAQTH----RAVDALTKCENDVNVIHAVARLFWRDKKIAKAR 196
+W + A FL + + TH RA+ +L + ++ V+VI A+L ++ +A+
Sbjct 1664 QEKSVWIKFATFLLKQGQGDGTHKLLQRALKSLPE-KDHVDVISKFAQLEFQLGDTERAK 1722
Query 197 KWMNRSVTLDPSLGDAWASYLAFELENGGEKECIDIINR 235
+++ P D W+ Y+ +++G +KE DI R
Sbjct 1723 ALFESTLSSYPKRTDLWSVYIDMMVKHGSQKEVRDIFER 1761
Score = 30.4 bits (67), Expect = 6.8, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 28/53 (52%), Gaps = 3/53 (5%)
Query 15 QLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAIDVF 67
QLE QLGD + A AL + + + LW V + +H +Q+++ D+F
Sbjct 1710 QLEFQLGDTERAKALFESTLSSYPKRTDLWSVYIDMMVKHGSQKEVR---DIF 1759
> dre:652952 ogt.2, im:7146393, ogtl, wu:fp46c04, wu:fr75f09;
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl
transferase)
2; K09667 polypeptide N-acetylglucosaminyltransferase [EC:2.4.1.-]
Length=1102
Score = 37.4 bits (85), Expect = 0.051, Method: Compositional matrix adjust.
Identities = 44/231 (19%), Positives = 88/231 (38%), Gaps = 14/231 (6%)
Query 4 INTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARA 63
IN NV ++ +R + Y A++L+ H A ++ +G + DLA
Sbjct 285 INLGNVLKEARIFDRAVAAYLRALSLSPNHAVVHGNLACVYYEQGLI--------DLA-- 334
Query 64 IDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEI 123
ID +R P + N + + N + A A P + L I+
Sbjct 335 IDTYRHAIELQPHFPDAYCNLANAMKEKCNVSEAEECYNTALRLCPTHADSLNNLANIKR 394
Query 124 ESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAV----DALTKCENDVNVI 179
E GN E A + KA++ P+ + + ++ + A+ +A+ +
Sbjct 395 EQGNIEEAVQLYRKALEVFPDFAAAHSNLASVLQQQGKLQEALMHYEEAIRISPTFADAY 454
Query 180 HAVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELENGGEKECI 230
+ + I A + R++ ++P+ DA ++ + ++G E I
Sbjct 455 SNMGNTLKEMQDIQGALRCYTRAIQINPAFADAHSNLASIHKDSGNIPEAI 505
> sce:YDR416W SYF1, NTC90; Member of the NineTeen Complex (NTC)
that contains Prp19p and stabilizes U6 snRNA in catalytic
forms of the spliceosome containing U2, U5, and U6 snRNAs; null
mutant has splicing defect and arrests in G2/M; homologs
in human and C. elegans; K12867 pre-mRNA-splicing factor SYF1
Length=859
Score = 37.4 bits (85), Expect = 0.063, Method: Composition-based stats.
Identities = 46/171 (26%), Positives = 77/171 (45%), Gaps = 34/171 (19%)
Query 1 RQNINTANVWMQSIQLERQLGD----YDAAIALAD-EAVKQHQTFAKLWMVRGQLHEEHP 55
RQ+ N WM+ + L++ + Y AI D V +F +LW G L+
Sbjct 402 RQDSNLVETWMKRVSLQKSAAEKCNVYSEAILKIDPRKVGTPGSFGRLWCSYGDLYWR-- 459
Query 56 TQRDLARAIDVFRE-GTVCCPRSVP-------LWLCWVNCERSQKNWNRARAVLEKAKLR 107
+ AI RE T P ++L W + E ++ RA ++LE A L
Sbjct 460 -----SNAISTARELWTQSLKVPYPYIEDLEEIYLNWADRELDKEGVERAFSILEDA-LH 513
Query 108 VPNSPELWLALVQIEIESGNREV-AQHVAAKAIQACPNAGIIWAEAI-FLE 156
VP +PE+ L + ++G+R++ AQ V +++ IW++ I +LE
Sbjct 514 VPTNPEILLE----KYKNGHRKIPAQTVLFNSLR-------IWSKYIDYLE 553
> pfa:PFD0180c CGI-201 protein, short form; K12869 crooked neck
Length=780
Score = 36.6 bits (83), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 22/87 (25%), Positives = 40/87 (45%), Gaps = 1/87 (1%)
Query 79 PLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKA 138
+++ + E Q N N+AR++ A +PN ++ + E++ GN ++V AK
Sbjct 410 KIFILYATFELRQLNVNKARSIFNNALQTIPNEK-IFEKFCEFELKLGNIRECRNVYAKY 468
Query 139 IQACPNAGIIWAEAIFLEEENAQTHRA 165
++A P W I E + RA
Sbjct 469 VEAFPFNSKAWISMINFELSLDEVERA 495
Score = 34.7 bits (78), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 19/70 (27%), Positives = 35/70 (50%), Gaps = 0/70 (0%)
Query 88 ERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQACPNAGI 147
E QK+ R R++ E+A + LWL +++E+ + N A+++ + + P I
Sbjct 81 EIKQKDIERCRSIFERALNIDYTNKNLWLKYIEVELINKNINSARNLLERVVLLLPLENI 140
Query 148 IWAEAIFLEE 157
W + LEE
Sbjct 141 FWKKYAHLEE 150
Score = 33.9 bits (76), Expect = 0.64, Method: Compositional matrix adjust.
Identities = 19/66 (28%), Positives = 29/66 (43%), Gaps = 0/66 (0%)
Query 57 QRDLARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWL 116
Q+D+ R +F + LWL ++ E KN N AR +LE+ L +P W
Sbjct 84 QKDIERCRSIFERALNIDYTNKNLWLKYIEVELINKNINSARNLLERVVLLLPLENIFWK 143
Query 117 ALVQIE 122
+E
Sbjct 144 KYAHLE 149
> dre:337685 ogt.1, fm81g08, ogt, wu:fc12b01, wu:fm81g08; O-linked
N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl
transferase) 1;
K09667 polypeptide N-acetylglucosaminyltransferase [EC:2.4.1.-]
Length=1062
Score = 36.6 bits (83), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 43/231 (18%), Positives = 87/231 (37%), Gaps = 14/231 (6%)
Query 4 INTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARA 63
IN NV ++ +R + Y A++L+ H A ++ +G + DLA
Sbjct 229 INLGNVLKEARIFDRAVAGYLRALSLSPNHAVVHGNLACVYYEQGLI--------DLA-- 278
Query 64 IDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEI 123
ID +R P + N + + N + A A P + L I+
Sbjct 279 IDTYRRAIELQPHFPDAYCNLANALKEKGNVSEAEECYNTALRLCPTHADSLNNLANIKR 338
Query 124 ESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAV----DALTKCENDVNVI 179
E GN E A + KA++ P + + ++ + A+ +A+ +
Sbjct 339 EQGNIEEAVQLYRKALEVFPEFAAAHSNLASVLQQQGKLQEALMHYKEAIRISPTFADAY 398
Query 180 HAVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFELENGGEKECI 230
+ + + A + R++ ++P+ DA ++ + ++G E I
Sbjct 399 SNMGNTLKEMQDVQGALQCYTRAIQINPAFADAHSNLASIHKDSGNIPEAI 449
> tpv:TP01_0087 adapter protein; K12867 pre-mRNA-splicing factor
SYF1
Length=839
Score = 36.6 bits (83), Expect = 0.10, Method: Composition-based stats.
Identities = 45/193 (23%), Positives = 76/193 (39%), Gaps = 28/193 (14%)
Query 1 RQNINTANVWMQSIQL-----ERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHP 55
+QNI+ W+ IQL R + Y A+ D + K +LW +EE
Sbjct 427 KQNIHNVYNWINYIQLFKDDPNRMVEIYAEAVQTIDVS-KSVGRVTELWSRFATFYEE-- 483
Query 56 TQRDLARAIDVFREGTVCCPRSV----PLWLCWVNCERSQKNWNRA-----RAVLEKAKL 106
+ DL A ++ + + + V LW CWV K + +A R+V K
Sbjct 484 -REDLENADKIYEKASNSDFKYVDDLATLWCCWVEMYLRHKQFKKALEISRRSVSGNGKT 542
Query 107 ----RVPNSPELWLALVQIEIESGNREVAQHVAAKAIQ---ACPNAGIIWAEAIFLEEEN 159
R+ +S +LW + +E G E + K ++ P + +A + E+N
Sbjct 543 SISRRLHSSVKLWTLALDMEQNFGTIETTRATFNKMVELKVVTPQVALSFAGYL---EQN 599
Query 160 AQTHRAVDALTKC 172
+ +A KC
Sbjct 600 KYFEASFNAFEKC 612
> dre:100150181 tmtc3; transmembrane and tetratricopeptide repeat
containing 3
Length=906
Score = 36.2 bits (82), Expect = 0.13, Method: Composition-based stats.
Identities = 53/226 (23%), Positives = 88/226 (38%), Gaps = 20/226 (8%)
Query 22 DYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAIDVFREGTVCCPRSVPLW 81
D+++ L A+K ++ AKLW G E Q RA+ F + T P +
Sbjct 427 DWESEYTLFTSALKVNRNNAKLWNNVGHALE---NQNSYERALRYFLQATRVQPDDIGAH 483
Query 82 LCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQA 141
+ N R+ KN NR+R E + P++ ++ A VA +
Sbjct 484 M---NVGRTYKNLNRSREAEEAYLIAKSLMPQI----------IPGKKYATRVAPNHLNV 530
Query 142 CPN-AGIIWAEAIFLEEENAQTHRAVDALTKCENDVNVIHAVARLFWRDKKIAKARKWMN 200
N A +I A LEE + Q +R A++ + + L + K ++A+
Sbjct 531 YINLANLIRANDSRLEEAD-QLYR--QAISMRPDFKQAYISRGELLLKMNKPSEAKDAYL 587
Query 201 RSVTLDPSLGDAWASYLAFELENGGEKECIDIINRCALAQPNRGLA 246
R++ LD + D W + +E E + NR P LA
Sbjct 588 RALELDHTNADLWYNLAIVNIEMKDPSEALRNFNRALDLNPRHKLA 633
> cpv:cgd2_3250 3x TPR domain-containing protein
Length=960
Score = 35.8 bits (81), Expect = 0.15, Method: Composition-based stats.
Identities = 33/126 (26%), Positives = 61/126 (48%), Gaps = 18/126 (14%)
Query 101 LEKAKLRVPNSPELWLAL----VQIEIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLE 156
L K +L+V +P L+ +L + + E+++H A+A ++ NA
Sbjct 625 LVKEQLKVRETPRLYCSLGDLTQDLSFYEKSWELSRHRFARAQRSLGNAYF--------- 675
Query 157 EENAQTHRAVDALTKCE----NDVNVIHAVARLFWRDKKIAKARKWMNRSVTLDPSLGDA 212
+ +Q A+DA TK +VN ++ + R ++ A++ +R V+LDP G+A
Sbjct 676 -KKSQFELALDAYTKASLVNSTNVNCWFSLGCVALRLERWEIAQQAFSRVVSLDPQQGEA 734
Query 213 WASYLA 218
WA+ A
Sbjct 735 WANLAA 740
> dre:393107 utp6, MGC174851, MGC55648, zgc:55648; UTP6, small
subunit (SSU) processome component, homolog (yeast); K14557
U3 small nucleolar RNA-associated protein 6
Length=594
Score = 35.8 bits (81), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 30/104 (28%), Positives = 47/104 (45%), Gaps = 9/104 (8%)
Query 66 VFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRV----PNSPELWLALVQI 121
VFR T V LWL V K W ++ L K L + P+ P LW+ +
Sbjct 95 VFRRATSKWQEDVQLWLSHVAF---NKKWG-SKTQLSKILLSMLAIHPDKPALWIMAAKC 150
Query 122 EIESGN-REVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHR 164
E+E N E A+H+ +A++ P ++ E +E +A+ R
Sbjct 151 EMEDRNSSESARHLFLRALRFHPENKKVYLEYFRMELMHAEKMR 194
> ath:AT4G24270 RNA recognition motif (RRM)-containing protein
Length=817
Score = 35.8 bits (81), Expect = 0.18, Method: Composition-based stats.
Identities = 29/141 (20%), Positives = 68/141 (48%), Gaps = 20/141 (14%)
Query 27 IALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAIDVFREGTVCCPRSVPLWLCWVN 86
+A+A++ +Q ++ R L EE+ +++DL+ + F+E ++ ++
Sbjct 262 VAVANKKAQQ------MYSERAHL-EENISKQDLSDT-EKFQE-----------FMNYIK 302
Query 87 CERSQKNWNRARAVLEKAKLRVPNSPELWLAL-VQIEIESGNREVAQHVAAKAIQACPNA 145
E++ + R +A+ E+A P S +LW+ V ++ + H ++A ++CP
Sbjct 303 FEKTSGDPTRVQAIYERAVAEYPVSSDLWIDYTVYLDKTLKVGKAITHAYSRATRSCPWT 362
Query 146 GIIWAEAIFLEEENAQTHRAV 166
G +WA + E + + + +
Sbjct 363 GDLWARYLLALERGSASEKEI 383
> mmu:217827 6720454P05Rik; cDNA sequence BC002230
Length=1167
Score = 35.4 bits (80), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 24/82 (29%), Positives = 43/82 (52%), Gaps = 6/82 (7%)
Query 95 NRARAVLEKAKLRVPNS---PELWLALVQIEIESGNREVAQHVAAKAIQACPNAGIIWAE 151
+R RA+ E A +R P LW + + GN+E ++ V KA+Q+CP A +++ +
Sbjct 1073 HRIRALFENA-IRSDKGNQCPLLWRMYLNFLVSLGNKERSKGVFYKALQSCPWAKVLYMD 1131
Query 152 AIFLEEENAQTHRAVDALTKCE 173
A +E + +D +T+ E
Sbjct 1132 A--MEYFPDELQEILDVMTEKE 1151
> pfa:PF14_0042 U3 small nucleolar ribonucleoprotein, U3 snoRNP,
putative; K14792 rRNA biogenesis protein RRP5
Length=468
Score = 35.0 bits (79), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 26/124 (20%), Positives = 53/124 (42%), Gaps = 6/124 (4%)
Query 5 NTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAI 64
N ++++ +I + ++ L +EA+K+ + K+W Q+ H T +D A
Sbjct 273 NEKSIYLHTIHILKKNKKLTQLKDLCEEAIKKFKYSKKIWSCYLQIL--HNTFKDEEYAH 330
Query 65 DVFREGTVCCPRSVPLWLCWVNCERSQKNWN---RARAVLEKAKLRVPNSPELWLALVQI 121
++ + P+ L + +N R + ++ R + EK P +LW + I
Sbjct 331 NILLKSLHSLPKKKHLNMI-INAARFEYKYSNKERGKTYFEKLIQEYPKRSDLWFTYLDI 389
Query 122 EIES 125
I S
Sbjct 390 HINS 393
> dre:368864 prpf39, fa10d07, si:dz261o22.3, wu:fa10d07; PRP39
pre-mRNA processing factor 39 homolog (yeast); K13217 pre-mRNA-processing
factor 39
Length=752
Score = 35.0 bits (79), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 30/139 (21%), Positives = 61/139 (43%), Gaps = 5/139 (3%)
Query 8 NVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAIDVF 67
N W + + E + G + + L + + + + W+ + E + T+ ++
Sbjct 429 NNWREYLDFELENGTPERVVVLFERCLIACALYEEFWIKYAKYLESYSTEA----VRHIY 484
Query 68 REG-TVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESG 126
++ TV P+ + L W E Q + + AR++L+ ++ VP + L V +E G
Sbjct 485 KKACTVHLPKKPNVHLLWAAFEEQQGSIDEARSILKAVEVSVPGLAMVRLRRVSLERRHG 544
Query 127 NREVAQHVAAKAIQACPNA 145
N E A+ + AI N+
Sbjct 545 NMEEAEALLQDAITNGRNS 563
Score = 32.0 bits (71), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 22/46 (47%), Gaps = 0/46 (0%)
Query 213 WASYLAFELENGGEKECIDIINRCALAQPNRGLAWNKTTKQVRCWS 258
W YL FELENG + + + RC +A W K K + +S
Sbjct 431 WREYLDFELENGTPERVVVLFERCLIACALYEEFWIKYAKYLESYS 476
Score = 30.4 bits (67), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 24/73 (32%), Positives = 32/73 (43%), Gaps = 9/73 (12%)
Query 63 AIDVFREGTVCCPRSVPLWLCWVNCERSQKNW------NRARAVLEKAKLRVPN---SPE 113
A +V+R G P SV LWL ++ R ++ +R RA E A L S
Sbjct 219 ADEVYRRGLQAIPLSVDLWLHYITFLRENQDTSDGEAESRIRASYEHAVLACGTDFRSDR 278
Query 114 LWLALVQIEIESG 126
LW A + E E G
Sbjct 279 LWEAYIAWETEQG 291
> hsa:160418 TMTC3, DKFZp686C0968, DKFZp686M1969, DKFZp686O22167,
DKFZp686O2342, FLJ90492, SMILE; transmembrane and tetratricopeptide
repeat containing 3
Length=914
Score = 35.0 bits (79), Expect = 0.28, Method: Composition-based stats.
Identities = 50/226 (22%), Positives = 89/226 (39%), Gaps = 20/226 (8%)
Query 22 DYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAIDVFREGTVCCPRSVPLW 81
D+++ L A+K ++ AKLW G E +++ RA+ F + T P +
Sbjct 427 DWESEYTLFMSALKVNKNNAKLWNNVGHALE---NEKNFERALKYFLQATHVQPDDIGAH 483
Query 82 LCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAIQA 141
+ N R+ KN NR + E + P++ ++ A +A +
Sbjct 484 M---NVGRTYKNLNRTKEAEESYMMAKSLMPQI----------IPGKKYAARIAPNHLNV 530
Query 142 CPN-AGIIWAEAIFLEEENAQTHRAVDALTKCENDVNVIHAVARLFWRDKKIAKARKWMN 200
N A +I A LEE + Q +R A++ + + L + K KA++
Sbjct 531 YINLANLIRANESRLEEAD-QLYR--QAISMRPDFKQAYISRGELLLKMNKPLKAKEAYL 587
Query 201 RSVTLDPSLGDAWASYLAFELENGGEKECIDIINRCALAQPNRGLA 246
+++ LD + D W + +E E + NR P LA
Sbjct 588 KALELDRNNADLWYNLAIVHIELKEPNEALKNFNRALELNPKHKLA 633
> hsa:3437 IFIT3, CIG-49, GARG-49, IFI60, IFIT4, IRG2, ISG60,
RIG-G; interferon-induced protein with tetratricopeptide repeats
3
Length=490
Score = 35.0 bits (79), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 42/169 (24%), Positives = 75/169 (44%), Gaps = 19/169 (11%)
Query 96 RARAVLEKAKLRVPNSPEL--WLALVQIEIESG-NREVAQHVAAKAIQACP-NAGIIWAE 151
RA+ EKA PN+PE LA+ +++ ++ + V +AI+ P N +
Sbjct 154 RAKVCFEKALEEKPNNPEFSSGLAIAMYHLDNHPEKQFSTDVLKQAIELSPDNQYVKVLL 213
Query 152 AIFLEEEN--AQTHRAV-DALTKCENDVNVIHAVARLFWRDKKIAKARKWMNRSVTLDPS 208
+ L++ N A+ + V +AL K +V+ + A+ + R + KA + R + P+
Sbjct 214 GLKLQKMNKEAEGEQFVEEALEKSPCQTDVLRSAAKFYRRKGDLDKAIELFQRVLESTPN 273
Query 209 LGDAWAS----YLA--------FELENGGEKECIDIINRCALAQPNRGL 245
G + Y A E E G KE I+ + + A+ N+ L
Sbjct 274 NGYLYHQIGCCYKAKVRQMQNTGESEASGNKEMIEALKQYAMDYSNKAL 322
> bbo:BBOV_III002230 17.m07215; tetratricopeptide repeat domain
containing protein; K09553 stress-induced-phosphoprotein 1
Length=546
Score = 35.0 bits (79), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 19/75 (25%), Positives = 39/75 (52%), Gaps = 3/75 (4%)
Query 1 RQNINTANVWMQSIQLERQLGDYDAAIALADEAVKQHQTFAKLWMVRGQLHEEHPTQRDL 60
R+N + ++ +LG+Y +A+A ++AV+ TF K W +G L H ++
Sbjct 388 RRNPSDIKLYTNRAAALTKLGEYPSALADCNKAVEMDPTFVKAWARKGNL---HVLLKEY 444
Query 61 ARAIDVFREGTVCCP 75
++A++ + +G P
Sbjct 445 SKALEAYDKGLALDP 459
> tgo:TGME49_069200 crooked neck-like protein 1, putative ; K12869
crooked neck
Length=794
Score = 35.0 bits (79), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 40/180 (22%), Positives = 74/180 (41%), Gaps = 19/180 (10%)
Query 60 LARAIDVFREGTVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPE--LWLA 117
L++ + V+ E P + W+ ++ E S+ + ++ R V E+A VP E W
Sbjct 436 LSKRVFVYEEELHGHPLNYDCWIDYIRLEESRGDIDKIRNVYERALANVPPVLEKRFWKR 495
Query 118 LVQI--------EIESGNREVAQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVD-- 167
V I E+++ + E + V K ++ P+ +A+ L R +D
Sbjct 496 YVYIWISYALFEELQAKDVERCRQVYVKMLEVIPHKKFSFAKIWSLYASFEVRQRDLDKA 555
Query 168 ------ALTKCENDVNVIHAVARLFWRDKKIAKARKWMNRSVTLDPSLGDAWASYLAFEL 221
A+ +C + A A+L R I + RK + + L P AW + + E+
Sbjct 556 RLIFGRAIAEC-GKPKIFVAYAQLELRLGCIDRCRKIYAKFIELHPFNPRAWIAMIDLEV 614
> dre:393920 crnkl1, MGC55327, zgc:55327; crooked neck pre-mRNA
splicing factor-like 1 (Drosophila); K12869 crooked neck
Length=753
Score = 34.7 bits (78), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 24/103 (23%), Positives = 48/103 (46%), Gaps = 8/103 (7%)
Query 39 TFAKLWMVRGQLHEEHPTQRDLARAIDVFREGTVC--CPRSVPLWLCWVNCERSQKNWNR 96
TFAK+W++ GQ ++ R + GT CP++ L+ ++ E + ++R
Sbjct 409 TFAKIWLLYGQFEIRQKNLQNARRGL-----GTAIGKCPKN-KLFKGYIELELQLREFDR 462
Query 97 ARAVLEKAKLRVPNSPELWLALVQIEIESGNREVAQHVAAKAI 139
R + EK P + W+ ++E G+ + ++ + AI
Sbjct 463 CRKLYEKYLEFSPENCTTWIKFAELETILGDTDRSRAIFELAI 505
Score = 34.3 bits (77), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 42/214 (19%), Positives = 78/214 (36%), Gaps = 36/214 (16%)
Query 12 QSIQLERQLGDYDAAIALA-DEAVKQHQTFAKLWMVRGQLHEEHPTQRDLARAIDVFREG 70
Q I + +L DY ++ +++++T W+ Q E + +++ R+ ++
Sbjct 50 QKITDKEELNDYKLKKRKGFEDNIRKNRTVISNWIKYAQWEE---SLQEVQRSRSIYERA 106
Query 71 TVCCPRSVPLWLCWVNCERSQKNWNRARAVLEKAKLRVPNSPELWLALVQIEIESGNREV 130
R++ LWL + E + N AR + ++A +P + W +E GN
Sbjct 107 LDVDHRNITLWLKYAEMEMKNRQVNHARNIWDRAITILPRVNQFWYKYTYMEEMLGN--- 163
Query 131 AQHVAAKAIQACPNAGIIWAEAIFLEEENAQTHRAVDALTKCENDVNVIHAVARLFWRDK 190
I C W E E + H+ R K
Sbjct 164 --------IAGCRQVFERWME--------------------WEPEEQAWHSYINFELRYK 195
Query 191 KIAKARKWMNRSVTLDPSLGDAWASYLAFELENG 224
++ KAR V + P + + W Y FE ++G
Sbjct 196 EVDKARSIYENFVMVHPEVKN-WIKYAHFEEKHG 228
> hsa:439996 IFIT1B, DKFZp781M1841, IFIT1L, bA149I23.6; interferon-induced
protein with tetratricopeptide repeats 1B; K14217
interferon-induced protein with tetratricopeptide repeats
1
Length=474
Score = 34.7 bits (78), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 30/129 (23%), Positives = 57/129 (44%), Gaps = 12/129 (9%)
Query 92 KNWNRARAVLEKAKLRVPNSPELWLA-------LVQIEIESG-NREVAQHVAAKAIQACP 143
KN+ RA+ EKA P +PE L + SG N+ + HV +A++ P
Sbjct 155 KNYERAKTCFEKALEGNPENPEFNTGYAITVYRLDKFNTASGRNKAFSLHVLKRAVRLNP 214
Query 144 N---AGIIWAEAIFLEEENAQTHRAV-DALTKCENDVNVIHAVARLFWRDKKIAKARKWM 199
+ ++ A + E + A+ + + +ALT + V A+ + R + KA + +
Sbjct 215 DDVYIRVLLALKLQDEGQEAEGEKYIEEALTSISSQAYVFQYAAKFYRRKGSVDKALELL 274
Query 200 NRSVTLDPS 208
++ P+
Sbjct 275 KMALETTPT 283
Lambda K H
0.320 0.131 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9724477856
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40