bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0606_orf3
Length=201
Score E
Sequences producing significant alignments: (Bits) Value
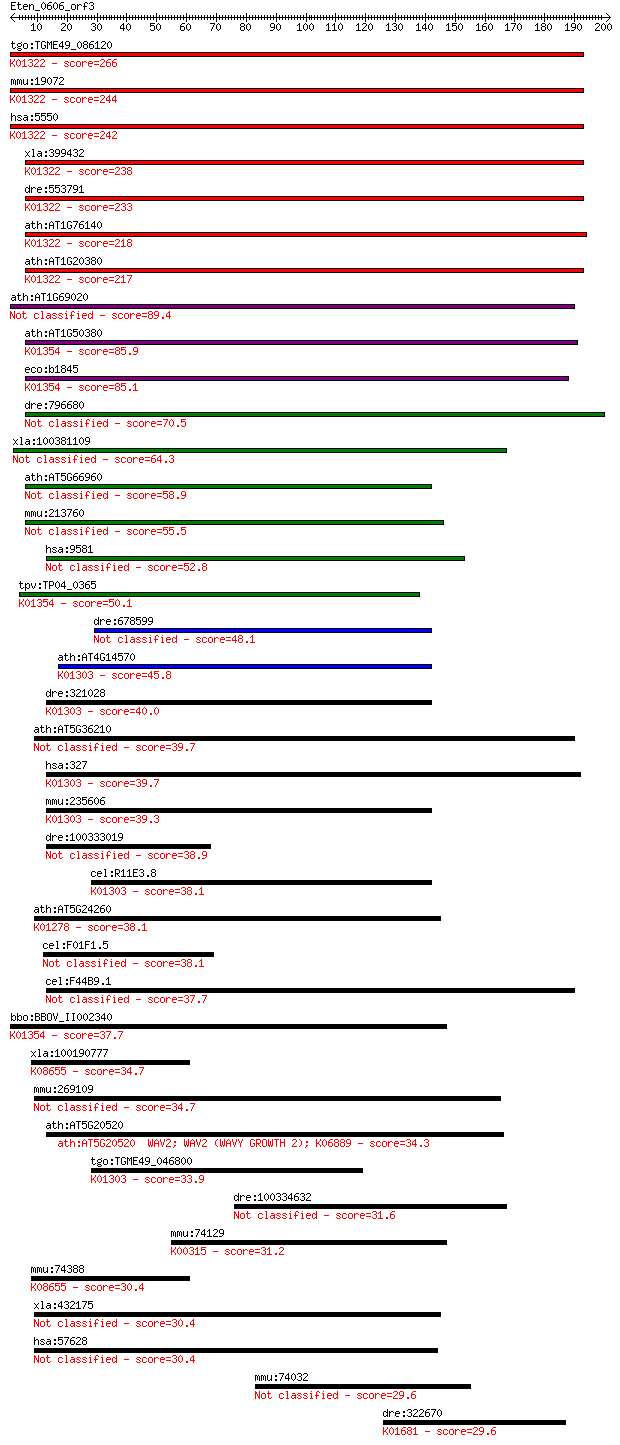
tgo:TGME49_086120 prolyl endopeptidase, putative (EC:3.4.21.26... 266 3e-71
mmu:19072 Prep, AI047692, AI450383, D10Wsu136e, PEP, Pop; prol... 244 1e-64
hsa:5550 PREP, MGC16060, PE, PEP; prolyl endopeptidase (EC:3.4... 242 7e-64
xla:399432 prep, xprep; prolyl endopeptidase (EC:3.4.21.26); K... 238 1e-62
dre:553791 prep, MGC110670, im:7140031, zgc:110670; prolyl end... 233 5e-61
ath:AT1G76140 serine-type endopeptidase/ serine-type peptidase... 218 2e-56
ath:AT1G20380 prolyl oligopeptidase, putative / prolyl endopep... 217 2e-56
ath:AT1G69020 prolyl oligopeptidase family protein 89.4 8e-18
ath:AT1G50380 prolyl oligopeptidase family protein; K01354 oli... 85.9 8e-17
eco:b1845 ptrB, ECK1846, JW1834, opdB, tlp; protease II (EC:3.... 85.1 1e-16
dre:796680 prepl, si:dkey-202m23.1; prolyl endopeptidase-like 70.5 3e-12
xla:100381109 prepl; prolyl endopeptidase-like (EC:3.4.21.-) 64.3 3e-10
ath:AT5G66960 prolyl oligopeptidase family protein 58.9 1e-08
mmu:213760 Prepl, 2810457N15Rik, 9530014L06Rik, D030028O16Rik,... 55.5 1e-07
hsa:9581 PREPL, FLJ16627, KIAA0436; prolyl endopeptidase-like ... 52.8 8e-07
tpv:TP04_0365 protease II (EC:3.4.21.83); K01354 oligopeptidas... 50.1 5e-06
dre:678599 MGC136971, si:dkey-16c7.3, wu:fc20g05; zgc:136971 48.1 2e-05
ath:AT4G14570 acylaminoacyl-peptidase-related; K01303 acylamin... 45.8 9e-05
dre:321028 apeh, cb5, sb:cb5, wu:fi37d02; acylpeptide hydrolas... 40.0 0.005
ath:AT5G36210 serine-type peptidase 39.7 0.006
hsa:327 APEH, ACPH, APH, D3F15S2, D3S48E, DNF15S2, MGC2178, OP... 39.7 0.007
mmu:235606 Apeh, MGC38101; acylpeptide hydrolase (EC:3.4.19.1)... 39.3 0.010
dre:100333019 N-acylaminoacyl-peptide hydrolase-like 38.9 0.012
cel:R11E3.8 dpf-5; Dipeptidyl Peptidase Four (IV) family membe... 38.1 0.021
ath:AT5G24260 prolyl oligopeptidase family protein; K01278 dip... 38.1 0.022
cel:F01F1.5 dpf-4; Dipeptidyl Peptidase Four (IV) family membe... 38.1 0.022
cel:F44B9.1 dpf-6; Dipeptidyl Peptidase Four (IV) family membe... 37.7 0.025
bbo:BBOV_II002340 18.m06190; hypothetical protein; K01354 olig... 37.7 0.026
xla:100190777 dpp8-a, dp8, dpp8, dpp8a, dprp1; dipeptidyl-pept... 34.7 0.25
mmu:269109 Dpp10, 6430601K09Rik, DPP_X, Dprp3; dipeptidylpepti... 34.7 0.25
ath:AT5G20520 WAV2; WAV2 (WAVY GROWTH 2); K06889 34.3 0.28
tgo:TGME49_046800 acylamino-acid-releasing enzyme, putative (E... 33.9 0.41
dre:100334632 dachsous 1-like 31.6 1.7
mmu:74129 Dmgdh, 1200014D15Rik, AI787269, MGC107623; dimethylg... 31.2 2.3
mmu:74388 Dpp8, 2310004I03Rik, 4932434F09Rik, AI666706, DPP_VI... 30.4 4.1
xla:432175 dpp10, MGC84485; dipeptidyl-peptidase 10 (non-funct... 30.4 4.3
hsa:57628 DPP10, DPL2, DPPY, DPRP3; dipeptidyl-peptidase 10 (n... 30.4 4.6
mmu:74032 Sdr42e1, 4632417N05Rik, AI117603; short chain dehydr... 29.6 7.6
dre:322670 aco2, cb1017, wu:fa10e03, wu:fb69g04, wu:fc20c11; a... 29.6 8.2
> tgo:TGME49_086120 prolyl endopeptidase, putative (EC:3.4.21.26);
K01322 prolyl oligopeptidase [EC:3.4.21.26]
Length=825
Score = 266 bits (680), Expect = 3e-71, Method: Compositional matrix adjust.
Identities = 127/194 (65%), Positives = 148/194 (76%), Gaps = 2/194 (1%)
Query 1 AAIKEKKQVSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVA 60
AAIK KKQVSYDDFQ AAEYL + Y+SP QLAI GGSNGGLLVGAC NQRPDL+ A +
Sbjct 629 AAIKTKKQVSYDDFQQAAEYLIAQKYTSPQQLAIEGGSNGGLLVGACINQRPDLYGAAII 688
Query 61 KVGVMDLLRFHKFTIGHAWVSDYGDPDKEEDFTHIYKISPLHNIKPEVAQSD--KQPAVL 118
VGV+DLLRFHKFTIGHAW SDYG+P+ EEDF I ISPLHNI + + PAVL
Sbjct 689 HVGVLDLLRFHKFTIGHAWTSDYGNPENEEDFPAILAISPLHNIGKGRGKGKGHQYPAVL 748
Query 119 VMTADHDDRVSPFHSLKYIAELQHAAGYSSPVSIPLIAHIDTKAGHGGGKPLMKVLEEQA 178
++T DHDDRVSPFHSLKYIAELQH+ G S + PL+ +DT GHG GKP+ K +EE A
Sbjct 749 LLTGDHDDRVSPFHSLKYIAELQHSVGSSPKQTNPLVIRVDTNTGHGAGKPVKKTIEEAA 808
Query 179 DTYGFIASVLGLKW 192
D YGF+A+ L ++W
Sbjct 809 DVYGFLANALHIQW 822
> mmu:19072 Prep, AI047692, AI450383, D10Wsu136e, PEP, Pop; prolyl
endopeptidase (EC:3.4.21.26); K01322 prolyl oligopeptidase
[EC:3.4.21.26]
Length=710
Score = 244 bits (624), Expect = 1e-64, Method: Compositional matrix adjust.
Identities = 114/192 (59%), Positives = 141/192 (73%), Gaps = 0/192 (0%)
Query 1 AAIKEKKQVSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVA 60
I KQ +DDFQ AAEYL K+GY+SP +L I GGSNGGLLV ACANQRPDLF V+A
Sbjct 517 GGILANKQNCFDDFQCAAEYLIKEGYTSPKRLTINGGSNGGLLVAACANQRPDLFGCVIA 576
Query 61 KVGVMDLLRFHKFTIGHAWVSDYGDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVM 120
+VGVMD+L+FHKFTIGHAW +DYG D ++ F + K SPLHN+K A + P++L++
Sbjct 577 QVGVMDMLKFHKFTIGHAWTTDYGCSDTKQHFEWLLKYSPLHNVKLPEADDIQYPSMLLL 636
Query 121 TADHDDRVSPFHSLKYIAELQHAAGYSSPVSIPLIAHIDTKAGHGGGKPLMKVLEEQADT 180
TADHDDRV P HSLK+IA LQ+ G S S PL+ H+DTKAGHG GKP KV+EE +D
Sbjct 637 TADHDDRVVPLHSLKFIATLQYIVGRSRKQSNPLLIHVDTKAGHGAGKPTAKVIEEVSDM 696
Query 181 YGFIASVLGLKW 192
+ FIA L ++W
Sbjct 697 FAFIARCLNIEW 708
> hsa:5550 PREP, MGC16060, PE, PEP; prolyl endopeptidase (EC:3.4.21.26);
K01322 prolyl oligopeptidase [EC:3.4.21.26]
Length=710
Score = 242 bits (617), Expect = 7e-64, Method: Compositional matrix adjust.
Identities = 113/192 (58%), Positives = 140/192 (72%), Gaps = 0/192 (0%)
Query 1 AAIKEKKQVSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVA 60
I KQ +DDFQ AAEYL K+GY+SP +L I GGSNGGLLV ACANQRPDLF V+A
Sbjct 517 GGILANKQNCFDDFQCAAEYLIKEGYTSPKRLTINGGSNGGLLVAACANQRPDLFGCVIA 576
Query 61 KVGVMDLLRFHKFTIGHAWVSDYGDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVM 120
+VGVMD+L+FHK+TIGHAW +DYG D ++ F + K SPLHN+K A + P++L++
Sbjct 577 QVGVMDMLKFHKYTIGHAWTTDYGCSDSKQHFEWLVKYSPLHNVKLPEADDIQYPSMLLL 636
Query 121 TADHDDRVSPFHSLKYIAELQHAAGYSSPVSIPLIAHIDTKAGHGGGKPLMKVLEEQADT 180
TADHDDRV P HSLK+IA LQ+ G S S PL+ H+DTKAGHG GKP KV+EE +D
Sbjct 637 TADHDDRVVPLHSLKFIATLQYIVGRSRKQSNPLLIHVDTKAGHGAGKPTAKVIEEVSDM 696
Query 181 YGFIASVLGLKW 192
+ FIA L + W
Sbjct 697 FAFIARCLNVDW 708
> xla:399432 prep, xprep; prolyl endopeptidase (EC:3.4.21.26);
K01322 prolyl oligopeptidase [EC:3.4.21.26]
Length=712
Score = 238 bits (606), Expect = 1e-62, Method: Compositional matrix adjust.
Identities = 110/187 (58%), Positives = 136/187 (72%), Gaps = 0/187 (0%)
Query 6 KKQVSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVM 65
KQ +DDFQ AAEYL K+GYSS + I GGSNGGLLV AC NQRPDLF +A+VGVM
Sbjct 524 NKQNCFDDFQCAAEYLVKEGYSSAKNITINGGSNGGLLVAACTNQRPDLFGCTIAQVGVM 583
Query 66 DLLRFHKFTIGHAWVSDYGDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTADHD 125
D+L+FHKFTIGHAW +D+G D +E F + K SPLHNI+ + P++L++TADHD
Sbjct 584 DMLKFHKFTIGHAWTTDFGCSDNKEHFDWLIKYSPLHNIRVPEKDGIQYPSMLLLTADHD 643
Query 126 DRVSPFHSLKYIAELQHAAGYSSPVSIPLIAHIDTKAGHGGGKPLMKVLEEQADTYGFIA 185
DRV P HSLK+IA LQ+ G S + PL+ H+DTKAGHG GKP KV+EE +D + FIA
Sbjct 644 DRVVPLHSLKFIASLQNIVGRSPNQTNPLLIHVDTKAGHGAGKPTAKVIEEVSDMFAFIA 703
Query 186 SVLGLKW 192
LGL+W
Sbjct 704 QCLGLQW 710
> dre:553791 prep, MGC110670, im:7140031, zgc:110670; prolyl endopeptidase
(EC:3.4.21.26); K01322 prolyl oligopeptidase [EC:3.4.21.26]
Length=709
Score = 233 bits (593), Expect = 5e-61, Method: Compositional matrix adjust.
Identities = 111/187 (59%), Positives = 132/187 (70%), Gaps = 0/187 (0%)
Query 6 KKQVSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVM 65
KQ + DFQ AAEYL K+GY+SP +L I GGSNGGLLV AC NQRPDLF VA+VGVM
Sbjct 521 NKQNCFTDFQCAAEYLIKEGYTSPKKLTINGGSNGGLLVAACVNQRPDLFGCAVAQVGVM 580
Query 66 DLLRFHKFTIGHAWVSDYGDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTADHD 125
D+L+FHKFTIGHAW +D+G + +E F + K SPLHNI+ + PAVL++T DHD
Sbjct 581 DMLKFHKFTIGHAWTTDFGCSEIKEQFDWLIKYSPLHNIQVPEGDGVQYPAVLLLTGDHD 640
Query 126 DRVSPFHSLKYIAELQHAAGYSSPVSIPLIAHIDTKAGHGGGKPLMKVLEEQADTYGFIA 185
DRV P HSLKYIA LQ+ G PL +IDTK+GHG GKP KV++E ADTY FIA
Sbjct 641 DRVVPLHSLKYIATLQNVIGQCPGQKNPLFIYIDTKSGHGAGKPTSKVIQEVADTYAFIA 700
Query 186 SVLGLKW 192
L L W
Sbjct 701 RCLNLSW 707
> ath:AT1G76140 serine-type endopeptidase/ serine-type peptidase;
K01322 prolyl oligopeptidase [EC:3.4.21.26]
Length=792
Score = 218 bits (554), Expect = 2e-56, Method: Compositional matrix adjust.
Identities = 104/192 (54%), Positives = 134/192 (69%), Gaps = 4/192 (2%)
Query 6 KKQVSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVM 65
KKQ +DDF S AEYL GY+ P +L I GGSNGGLLVGAC NQRPDL+ +A VGVM
Sbjct 600 KKQNCFDDFISGAEYLVSAGYTQPSKLCIEGGSNGGLLVGACINQRPDLYGCALAHVGVM 659
Query 66 DLLRFHKFTIGHAWVSDYGDPDKEEDFTHIYKISPLHNIK-PEVAQSD---KQPAVLVMT 121
D+LRFHKFTIGHAW SDYG + EE+F + K SPLHN+K P Q+D + P+ +++T
Sbjct 660 DMLRFHKFTIGHAWTSDYGCSENEEEFHWLIKYSPLHNVKRPWEQQTDHLVQYPSTMLLT 719
Query 122 ADHDDRVSPFHSLKYIAELQHAAGYSSPVSIPLIAHIDTKAGHGGGKPLMKVLEEQADTY 181
ADHDDRV P HSLK +A + + +SP P+I I+ KAGHG G+P K+++E AD Y
Sbjct 720 ADHDDRVVPLHSLKLLAHVLCTSLDNSPQMNPIIGRIEVKAGHGAGRPTQKMIDEAADRY 779
Query 182 GFIASVLGLKWS 193
F+A ++ W+
Sbjct 780 SFMAKMVNASWT 791
> ath:AT1G20380 prolyl oligopeptidase, putative / prolyl endopeptidase,
putative / post-proline cleaving enzyme, putative;
K01322 prolyl oligopeptidase [EC:3.4.21.26]
Length=731
Score = 217 bits (552), Expect = 2e-56, Method: Compositional matrix adjust.
Identities = 105/194 (54%), Positives = 136/194 (70%), Gaps = 7/194 (3%)
Query 6 KKQVSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVM 65
KQ +DDF S AEYL GY+ P +L I GGSNGG+LVGAC NQRPDLF +A VGVM
Sbjct 536 NKQNCFDDFISGAEYLVSAGYTQPRKLCIEGGSNGGILVGACINQRPDLFGCALAHVGVM 595
Query 66 DLLRFHKFTIGHAWVSDYGDPDKEEDFTHIYKISPLHNIK-PEVAQSD---KQPAVLVMT 121
D+LRFHKFTIGHAW S++G DKEE+F + K SPLHN+K P ++D + P+ +++T
Sbjct 596 DMLRFHKFTIGHAWTSEFGCSDKEEEFHWLIKYSPLHNVKRPWEQKTDLFFQYPSTMLLT 655
Query 122 ADHDDRVSPFHSLKYIAELQHAAGYS---SPVSIPLIAHIDTKAGHGGGKPLMKVLEEQA 178
ADHDDRV P HS K +A +Q+ G S SP + P+IA I+ KAGHG G+P K+++E A
Sbjct 656 ADHDDRVVPLHSYKLLATMQYELGLSLENSPQTNPIIARIEVKAGHGAGRPTQKMIDEAA 715
Query 179 DTYGFIASVLGLKW 192
D Y F+A ++ W
Sbjct 716 DRYSFMAKMVDASW 729
> ath:AT1G69020 prolyl oligopeptidase family protein
Length=757
Score = 89.4 bits (220), Expect = 8e-18, Method: Compositional matrix adjust.
Identities = 61/195 (31%), Positives = 94/195 (48%), Gaps = 15/195 (7%)
Query 1 AAIKEKKQVSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVA 60
+ + KQ S DF +A+YL +KGY LA +G S G +L A N P LF+AV+
Sbjct 567 SGTRSLKQNSIQDFIYSAKYLVEKGYVHRHHLAAVGYSAGAILPAAAMNMHPSLFQAVIL 626
Query 61 KVGVMDLL------RFHKFTIGHAWVSDYGDPDKEEDFTHIYKISPLHNIKPEVAQSDKQ 114
KV +D+L + H ++G+PD + DF I SP I+ +V
Sbjct 627 KVPFVDVLNTLSDPNLPLTLLDH---EEFGNPDNQTDFGSILSYSPYDKIRKDVC----Y 679
Query 115 PAVLVMTADHDDRVSPFHSLKYIAELQHAAGYSSPVSIPLIAHIDTKAGHGGGKPLMKVL 174
P++LV T+ HD RV + K++A+++ + + S +I + GH G
Sbjct 680 PSMLVTTSFHDSRVGVWEGAKWVAKIRDSTCHD--CSRAVILKTNMNGGHFGEGGRYAQC 737
Query 175 EEQADTYGFIASVLG 189
EE A Y F+ V+G
Sbjct 738 EETAFDYAFLLKVMG 752
> ath:AT1G50380 prolyl oligopeptidase family protein; K01354 oligopeptidase
B [EC:3.4.21.83]
Length=710
Score = 85.9 bits (211), Expect = 8e-17, Method: Compositional matrix adjust.
Identities = 60/188 (31%), Positives = 98/188 (52%), Gaps = 12/188 (6%)
Query 6 KKQVSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVM 65
KK+ ++ DF + AE L + Y S ++L + G S GGLL+GA N RPDLF+ V+A V +
Sbjct 527 KKKNTFTDFIACAERLIELKYCSKEKLCMEGRSAGGLLMGAVVNMRPDLFKVVIAGVPFV 586
Query 66 DLLRFH---KFTIGHAWVSDYGDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTA 122
D+L + + ++GDP KEE + ++ SP+ N+ AQ+ P +LV
Sbjct 587 DVLTTMLDPTIPLTTSEWEEWGDPRKEEFYFYMKSYSPVDNV---TAQN--YPNMLVTAG 641
Query 123 DHDDRVSPFHSLKYIAELQHAAGYSSPVSIPLIAHIDTKAGHGGGKPLMKVLEEQADTYG 182
+D RV K++A+L+ ++ L+ + AGH + L+E A T+
Sbjct 642 LNDPRVMYSEPGKWVAKLREMKTDNN----VLLFKCELGAGHFSKSGRFEKLQEDAFTFA 697
Query 183 FIASVLGL 190
F+ VL +
Sbjct 698 FMMKVLDM 705
> eco:b1845 ptrB, ECK1846, JW1834, opdB, tlp; protease II (EC:3.4.21.83);
K01354 oligopeptidase B [EC:3.4.21.83]
Length=686
Score = 85.1 bits (209), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 56/187 (29%), Positives = 91/187 (48%), Gaps = 16/187 (8%)
Query 6 KKQVSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVM 65
KK+ +++D+ A + L K GY SP MGGS GG+L+G NQRP+LF V+A+V +
Sbjct 500 KKKNTFNDYLDACDALLKLGYGSPSLCYAMGGSAGGMLMGVAINQRPELFHGVIAQVPFV 559
Query 66 DLLRFH-----KFTIGHAWVSDYGDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVM 120
D++ T G ++G+P + + ++ SP N+ + P +LV
Sbjct 560 DVVTTMLDESIPLTTGE--FEEWGNPQDPQYYEYMKSYSPYDNVTAQA-----YPHLLVT 612
Query 121 TADHDDRVSPFHSLKYIAELQHAAGYSSPVSIPLIAHIDTKAGHGGGKPLMKVLEEQADT 180
T HD +V + K++A+L+ L+ D +GHGG K E A
Sbjct 613 TGLHDSQVQYWEPAKWVAKLRELKTDDH----LLLLCTDMDSGHGGKSGRFKSYEGVAME 668
Query 181 YGFIASV 187
Y F+ ++
Sbjct 669 YAFLVAL 675
> dre:796680 prepl, si:dkey-202m23.1; prolyl endopeptidase-like
Length=692
Score = 70.5 bits (171), Expect = 3e-12, Method: Composition-based stats.
Identities = 55/203 (27%), Positives = 89/203 (43%), Gaps = 14/203 (6%)
Query 6 KKQVSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVM 65
+KQ +D + + L + G S PD A+ S G +L GA NQ P L RA++ + +
Sbjct 492 QKQRGVEDLAACIQTLHRLGVSGPDLTALTARSAGAILAGALCNQNPQLIRALILQAPFL 551
Query 66 DLLRFHKFT---IGHAWVSDYGDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTA 122
D+L + T + ++GDP E +I P HNI+P++ P++L+
Sbjct 552 DVLGTMQDTSLPLTLEEKGEWGDPLIREHRDNIASYCPCHNIQPQL-----YPSMLITAY 606
Query 123 DHDDRVSPFHSLKYIAELQHAAGYSSP-----VSIP-LIAHIDTKAGHGGGKPLMKVLEE 176
D RV +K++ L+ A + +P +I I A H G + L E
Sbjct 607 SEDHRVPVSGVIKFVERLKKAIQMCTTQVDINARVPSVILDIQPGADHFGPEDFHLSLNE 666
Query 177 QADTYGFIASVLGLKWSCPETEK 199
A F+ + LGL T +
Sbjct 667 SARQLAFLYTELGLSLDQQRTRR 689
> xla:100381109 prepl; prolyl endopeptidase-like (EC:3.4.21.-)
Length=707
Score = 64.3 bits (155), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 54/174 (31%), Positives = 75/174 (43%), Gaps = 16/174 (9%)
Query 2 AIKEKKQVSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAK 61
+ +KK +D S +L GYS P A+ S GG+L GA N P LFRAVV +
Sbjct 502 GVLDKKLNGLEDLGSCISHLHGLGYSQPHYSAVEAASAGGVLAGALCNSAPRLFRAVVLE 561
Query 62 VGVMDLLRFHK-----FTIGHAWVSDYGDPDKEEDF-THIYKISPLHNIKPEVAQSDKQP 115
+D+L TI ++G+P +E + +I P NI P+ P
Sbjct 562 APFLDVLNTMMNVSLPLTIEEQ--EEWGNPLSDEKYHRYIKSYCPYQNITPQ-----NYP 614
Query 116 AVLVMTADHDDRVSPFHSLKYIAELQHAA---GYSSPVSIPLIAHIDTKAGHGG 166
V + ++D RV L YI L+ AA + S S I HI GG
Sbjct 615 CVRITAYENDQRVPIQGLLGYITRLRKAARDYCHESGTSESRIPHIYLDVHPGG 668
> ath:AT5G66960 prolyl oligopeptidase family protein
Length=792
Score = 58.9 bits (141), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 47/139 (33%), Positives = 67/139 (48%), Gaps = 8/139 (5%)
Query 6 KKQVSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVM 65
KK S D+ A+YL + ++LA G S GGL+V + N PDLF+A V KV +
Sbjct 615 KKLNSIKDYIQCAKYLVENNIVEENKLAGWGYSAGGLVVASAINHCPDLFQAAVLKVPFL 674
Query 66 DLLRFHKFTIGHAWVSDY---GDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTA 122
D + I DY G P DF I + SP NI +V PAVLV T+
Sbjct 675 DPTHTLIYPILPLTAEDYEEFGYPGDINDFHAIREYSPYDNIPKDVL----YPAVLV-TS 729
Query 123 DHDDRVSPFHSLKYIAELQ 141
+ R + + K++A ++
Sbjct 730 SFNTRFGVWEAAKWVARVR 748
> mmu:213760 Prepl, 2810457N15Rik, 9530014L06Rik, D030028O16Rik,
MGC7980, mKIAA0436; prolyl endopeptidase-like (EC:3.4.21.-)
Length=725
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 39/144 (27%), Positives = 67/144 (46%), Gaps = 9/144 (6%)
Query 6 KKQVSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVM 65
KK D + + L +G+S P + S GG+LVGA N +P+L RAV + +
Sbjct 525 KKLNGLADLVACIKTLHSQGFSQPSLTTLSAFSAGGVLVGALCNSKPELLRAVTLEAPFL 584
Query 66 DLLRF---HKFTIGHAWVSDYGDPDKEEDF-THIYKISPLHNIKPEVAQSDKQPAVLVMT 121
D+L + + ++G+P +E +I + P NIKP+ P+V +
Sbjct 585 DVLNTMLDTTLPLTLEELEEWGNPSSDEKHKNYIKRYCPCQNIKPQ-----HYPSVHITA 639
Query 122 ADHDDRVSPFHSLKYIAELQHAAG 145
++D+RV + Y +L+ A
Sbjct 640 YENDERVPLKGIVNYTEKLKEAVA 663
> hsa:9581 PREPL, FLJ16627, KIAA0436; prolyl endopeptidase-like
(EC:3.4.21.-)
Length=665
Score = 52.8 bits (125), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 39/153 (25%), Positives = 69/153 (45%), Gaps = 18/153 (11%)
Query 13 DFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLLRF-- 70
D ++ + L +G+S P + S GG+L GA N P+L RAV + +D+L
Sbjct 472 DLEACIKTLHGQGFSQPSLTTLTAFSAGGVLAGALCNSNPELVRAVTLEAPFLDVLNTMM 531
Query 71 -HKFTIGHAWVSDYGDPDKEEDF-THIYKISPLHNIKPEVAQSDKQPAVLVMTADHDDRV 128
+ + ++G+P +E +I + P NIKP+ P++ + ++D+RV
Sbjct 532 DTTLPLTLEELEEWGNPSSDEKHKNYIKRYCPYQNIKPQ-----HYPSIHITAYENDERV 586
Query 129 SPFHSLKYIAELQHA---------AGYSSPVSI 152
+ Y +L+ A GY +P I
Sbjct 587 PLKGIVSYTEKLKEAIAEHAKDTGEGYQTPNII 619
> tpv:TP04_0365 protease II (EC:3.4.21.83); K01354 oligopeptidase
B [EC:3.4.21.83]
Length=767
Score = 50.1 bits (118), Expect = 5e-06, Method: Composition-based stats.
Identities = 39/145 (26%), Positives = 65/145 (44%), Gaps = 18/145 (12%)
Query 4 KEKKQVSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVG 63
K+KK S+ D ++L K +S +LAI S G+L G N RPDL + K+
Sbjct 583 KDKKYNSFYDLVDVIQFLISKNVTSRSKLAINVSSASGILGGCVYNMRPDLCSVISFKLP 642
Query 64 VMDLLRFHKFTIG-------HAWVSDYGDPD----KEEDFTHIYKISPLHNIKPEVAQSD 112
+D H T+ ++G +E + HIY + P N+K
Sbjct 643 YLD----HFATVNDPSQPLVELEYEEFGSTQEYECEEYNVDHIYSMDPCVNMK---KVHS 695
Query 113 KQPAVLVMTADHDDRVSPFHSLKYI 137
K+P++L+ +D R +H+ K++
Sbjct 696 KRPSILINCNINDVRAPWYHAAKFL 720
> dre:678599 MGC136971, si:dkey-16c7.3, wu:fc20g05; zgc:136971
Length=714
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 34/122 (27%), Positives = 58/122 (47%), Gaps = 17/122 (13%)
Query 29 PDQLAIMGGSNGGLLVGACANQRPDLFRAVVAK---------VGVMDLLRFHKFTIGHAW 79
PD++A+MGGS+GG L Q PD +RA A+ +G D++ + ++G +
Sbjct 561 PDRVAVMGGSHGGFLACHLVGQYPDFYRACAARNPVINAATLLGTSDIVDWRYSSVGLQY 620
Query 80 VSDYGDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTADHDDRVSPFHSLKYIAE 139
D P + + + K +H P++ + VL+M + D RVSP L+
Sbjct 621 AFDR-LPTSQSLISMLDKSPIIH--APQI-----RAPVLLMLGERDRRVSPHQGLELYRA 672
Query 140 LQ 141
L+
Sbjct 673 LK 674
> ath:AT4G14570 acylaminoacyl-peptidase-related; K01303 acylaminoacyl-peptidase
[EC:3.4.19.1]
Length=764
Score = 45.8 bits (107), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 32/133 (24%), Positives = 57/133 (42%), Gaps = 14/133 (10%)
Query 17 AAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLLRFHKFTIG 76
A ++ + G + P ++ ++GGS+GG L Q PD F A A+ V ++ T
Sbjct 597 AVDHAIEMGIADPSRITVLGGSHGGFLTTHLIGQAPDKFVAAAARNPVCNMASMVGITDI 656
Query 77 HAWV--------SDYGDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTADHDDRV 128
W S Y + ED + +++SP+ +I S + L + D RV
Sbjct 657 PDWCFFEAYGDQSHYTEAPSAEDLSRFHQMSPISHI------SKVKTPTLFLLGTKDLRV 710
Query 129 SPFHSLKYIAELQ 141
+ +Y+ L+
Sbjct 711 PISNGFQYVRALK 723
> dre:321028 apeh, cb5, sb:cb5, wu:fi37d02; acylpeptide hydrolase
(EC:3.4.19.1); K01303 acylaminoacyl-peptidase [EC:3.4.19.1]
Length=730
Score = 40.0 bits (92), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 35/130 (26%), Positives = 55/130 (42%), Gaps = 1/130 (0%)
Query 13 DFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLLRFHK 72
D Q A + + K+G ++A++GGS+GG L Q P ++A VA+ V +L
Sbjct 560 DVQFAVDSVLKQGGFDEQKVAVIGGSHGGFLACHLIGQYPGFYKACVARNPVTNLASMVC 619
Query 73 FTIGHAWVSDYGDPDKEEDFTHIYKISPLHNIKPEVAQSDK-QPAVLVMTADHDDRVSPF 131
T W D + D I IK + K + VL+M + D RV
Sbjct 620 CTDIPDWCIVEAGFDYKPDIQLEPAILEQMLIKSPIKHVAKVKTPVLLMLGEGDKRVPNK 679
Query 132 HSLKYIAELQ 141
++Y L+
Sbjct 680 QGIEYYKALK 689
> ath:AT5G36210 serine-type peptidase
Length=730
Score = 39.7 bits (91), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 51/184 (27%), Positives = 80/184 (43%), Gaps = 24/184 (13%)
Query 9 VSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLL 68
V DD A+YL G + +L I GGS GG A R D+F+A + GV DL
Sbjct 550 VDVDDCCGCAKYLVSSGKADVKRLCISGGSAGGYTTLASLAFR-DVFKAGASLYGVADLK 608
Query 69 RFHKFTIGHAWVSDYGDP--DKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTADHDD 126
+ GH + S Y D E+DF Y+ SP++ + DK +++ +D
Sbjct 609 MLKE--EGHKFESRYIDNLVGDEKDF---YERSPINFV-------DKFSCPIILFQGLED 656
Query 127 R-VSPFHSLKYIAELQHAAGYSSPVSIPLIAHIDTKAGHGGGKPLMKVLEEQADTYGFIA 185
+ V+P S K L+ + + L+ + + G + + LE+Q F A
Sbjct 657 KVVTPDQSRKIYEALKKKG-----LPVALVEYEGEQHGFRKAENIKYTLEQQM---VFFA 708
Query 186 SVLG 189
V+G
Sbjct 709 RVVG 712
> hsa:327 APEH, ACPH, APH, D3F15S2, D3S48E, DNF15S2, MGC2178,
OPH; N-acylaminoacyl-peptide hydrolase (EC:3.4.19.1); K01303
acylaminoacyl-peptidase [EC:3.4.19.1]
Length=732
Score = 39.7 bits (91), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 40/180 (22%), Positives = 79/180 (43%), Gaps = 15/180 (8%)
Query 13 DFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLLRFHK 72
D Q A E + ++ + +A+MGGS+GG + Q P+ +RA VA+ V+++
Sbjct 562 DVQFAVEQVLQEEHFDASHVALMGGSHGGFISCHLIGQYPETYRACVARNPVINIASMLG 621
Query 73 FTIGHAW-VSDYGDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTADHDDRVSPF 131
T W V + G P + + + + + P + +L+M D RV
Sbjct 622 STDIPDWCVVEAGFPFSSDCLPDLSVWAEMLDKSPIRYIPQVKTPLLLMLGQEDRRVPFK 681
Query 132 HSLKYIAELQHAAGYSSPVSIPLIAHIDTKAGHGGGKPLMKVLEEQADTYGFIASVLGLK 191
++Y L+ ++P+ + K+ H + +E ++D+ F+ +VL L+
Sbjct 682 QGMEYYRALKTR-------NVPVRLLLYPKSTHA-----LSEVEVESDS--FMNAVLWLR 727
> mmu:235606 Apeh, MGC38101; acylpeptide hydrolase (EC:3.4.19.1);
K01303 acylaminoacyl-peptidase [EC:3.4.19.1]
Length=732
Score = 39.3 bits (90), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 30/130 (23%), Positives = 58/130 (44%), Gaps = 1/130 (0%)
Query 13 DFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLLRFHK 72
D Q A + + ++ + ++A+MGGS+GG L Q P+ + A +A+ V++++
Sbjct 562 DVQFAVQQVLQEEHFDARRVALMGGSHGGFLSCHLIGQYPETYSACIARNPVINIVSMMG 621
Query 73 FTIGHAW-VSDYGDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTADHDDRVSPF 131
T W + + G P + + + + + P + VL+M D RV
Sbjct 622 TTDIPDWCMVETGFPYSNDYLPDLNVLEEMLDKSPIKYIPQVKTPVLLMLGQEDRRVPFK 681
Query 132 HSLKYIAELQ 141
L+Y L+
Sbjct 682 QGLEYYHALK 691
> dre:100333019 N-acylaminoacyl-peptide hydrolase-like
Length=742
Score = 38.9 bits (89), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 19/55 (34%), Positives = 32/55 (58%), Gaps = 0/55 (0%)
Query 13 DFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDL 67
D Q A + + K+G ++A++GGS+GG L Q P ++A VA+ V++L
Sbjct 572 DVQFAVDSVLKQGGFDEQKVAVIGGSHGGFLACHLIGQYPGFYKACVARNPVINL 626
> cel:R11E3.8 dpf-5; Dipeptidyl Peptidase Four (IV) family member
(dpf-5); K01303 acylaminoacyl-peptidase [EC:3.4.19.1]
Length=737
Score = 38.1 bits (87), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 31/119 (26%), Positives = 50/119 (42%), Gaps = 9/119 (7%)
Query 28 SPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLLRFHKFTIGHAW-----VSD 82
S D++ + GGS+GG LV Q P +++ VA V+++ H T W +
Sbjct 582 SRDKVVLFGGSHGGFLVSHLIGQYPGFYKSCVALNPVVNIATMHDITDIPEWCYFEGTGE 641
Query 83 YGDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTADHDDRVSPFHSLKYIAELQ 141
Y D K T + + N P + L++ + D RV P H +I L+
Sbjct 642 YPDWTK---ITTTEQREKMFNSSPIAHVENATTPYLLLIGEKDLRVVP-HYRAFIRALK 696
> ath:AT5G24260 prolyl oligopeptidase family protein; K01278 dipeptidyl-peptidase
4 [EC:3.4.14.5]
Length=746
Score = 38.1 bits (87), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 35/137 (25%), Positives = 61/137 (44%), Gaps = 13/137 (9%)
Query 9 VSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLL 68
V +D + A++L ++G + PD + + G S GG L + P++F V+ V
Sbjct 586 VDAEDQVTGAKWLIEQGLAKPDHIGVYGWSYGGYLSATLLTRYPEIFNCAVSGAPVTSWD 645
Query 69 RFHKFTIGHAWVSDY-GDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTADHDDR 127
+ F + Y G P +EE + K S +H++ +DKQ +LV D+
Sbjct 646 GYDSF-----YTEKYMGLPTEEERY---LKSSVMHHVG---NLTDKQKLMLVHGM-IDEN 693
Query 128 VSPFHSLKYIAELQHAA 144
V H+ + + L A
Sbjct 694 VHFRHTARLVNALVEAG 710
> cel:F01F1.5 dpf-4; Dipeptidyl Peptidase Four (IV) family member
(dpf-4)
Length=629
Score = 38.1 bits (87), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 33/57 (57%), Gaps = 0/57 (0%)
Query 12 DDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLL 68
DD + A+ L ++G +++ I G S+GG L+ +C ++ +A V+ GV DLL
Sbjct 449 DDMLNGAKALVEQGKVDAEKVLITGSSSGGYLILSCLISPKNIIKAAVSVYGVADLL 505
> cel:F44B9.1 dpf-6; Dipeptidyl Peptidase Four (IV) family member
(dpf-6)
Length=740
Score = 37.7 bits (86), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 46/185 (24%), Positives = 72/185 (38%), Gaps = 23/185 (12%)
Query 13 DFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLLRFHK 72
D A E+ KG ++ ++A+MGGS GG P F V VG +L+ +
Sbjct 491 DILDAVEFAVSKGIANRSEVAVMGGSYGGYETLVALTFTPQTFACGVDIVGPSNLISLVQ 550
Query 73 FTIGHAWVSDYGDPDK--------EEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTADH 124
I W+ D K EE + SPL A +P ++++ +
Sbjct 551 -AIPPYWLGFRKDLIKMVGADISDEEGRQSLQSRSPLF-----FADRVTKP-IMIIQGAN 603
Query 125 DDRVSPFHSLKYIAELQHAAGYSSPVSIPLIAHIDTKAGHGGGKPLMKVLEEQADTYGFI 184
D RV S +++A L+ IP+ + GHG KP +E+ F+
Sbjct 604 DPRVKQAESDQFVAALEKK-------HIPVTYLLYPDEGHGVRKP-QNSMEQHGHIETFL 655
Query 185 ASVLG 189
LG
Sbjct 656 QQCLG 660
> bbo:BBOV_II002340 18.m06190; hypothetical protein; K01354 oligopeptidase
B [EC:3.4.21.83]
Length=487
Score = 37.7 bits (86), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 34/155 (21%), Positives = 57/155 (36%), Gaps = 23/155 (14%)
Query 1 AAIKEKKQVSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVA 60
A+K K D E+L + +A+ S G +L N RPDL +
Sbjct 306 GAVKSFKYKGVYDLIDVIEFLIANNMAQRSSIALSSTSAGAILAACVYNMRPDLCNCISL 365
Query 61 KVGVMDLLRFHKFTIGHAWVSDYGDP----DKEE-----DFTHIYKISPLHNIKPEVAQS 111
K+ +D+ + D +P +KEE + +Y SP N
Sbjct 366 KLPFLDVF---------GSIHDTNEPLSSLEKEEFGNESNVECVYSYSPCDNTG---EPG 413
Query 112 DKQPAVLVMTADHDDRVSPFHSLKYIAELQHAAGY 146
+P++++ D R H++ YI QH +
Sbjct 414 HLKPSLIIQCNSDDTRAPLRHTVSYIR--QHRTNW 446
> xla:100190777 dpp8-a, dp8, dpp8, dpp8a, dprp1; dipeptidyl-peptidase
8; K08655 dipeptidyl-peptidase 8 [EC:3.4.14.5]
Length=888
Score = 34.7 bits (78), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 20/54 (37%), Positives = 27/54 (50%), Gaps = 1/54 (1%)
Query 8 QVSYDDFQSAAEYLF-KKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVA 60
QV DD +YL K + D++ + G S GG L QRPD+FR +A
Sbjct 709 QVEIDDQVEGLQYLAAKHSFIDLDRVGVHGWSYGGYLSLMALVQRPDIFRVAIA 762
> mmu:269109 Dpp10, 6430601K09Rik, DPP_X, Dprp3; dipeptidylpeptidase
10
Length=800
Score = 34.7 bits (78), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 47/161 (29%), Positives = 65/161 (40%), Gaps = 20/161 (12%)
Query 9 VSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFR--AVVAKVGVMD 66
V D +A +YL K+ Y +L+I G GG + F+ AVVA + M
Sbjct 626 VEAKDQVAAVKYLLKQPYIDSKRLSIFGKGYGGYIASMILKSDEKFFKCGAVVAPISDMK 685
Query 67 LLRFHKFTIGHAWVSDY-GDPDKEEDFTHIYKISP-LHNIKPEVAQSDKQPAVLVMTADH 124
L A+ Y G P KEE Y+ S LHNI K+ +L++
Sbjct 686 LY-------ASAFSERYLGMPSKEES---TYQASSVLHNI-----HGLKEENLLIIHGTA 730
Query 125 DDRVSPFHSLKYIAELQHA-AGYSSPVSIPLIAHIDTKAGH 164
D +V HS + I L A Y+ V HI K+ H
Sbjct 731 DTKVHFQHSAELIKHLIKAGVNYTLQVYPDEGYHISDKSKH 771
> ath:AT5G20520 WAV2; WAV2 (WAVY GROWTH 2); K06889
Length=308
Score = 34.3 bits (77), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 40/160 (25%), Positives = 62/160 (38%), Gaps = 17/160 (10%)
Query 13 DFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAK---VGVMD--- 66
D Q+A ++L + ++ + G S GG + PD A++ + ++D
Sbjct 134 DAQAALDHLSGRTDIDTSRIVVFGRSLGGAVGAVLTKNNPDKVSALILENTFTSILDMAG 193
Query 67 -LLRFHKFTIGHAWVSDYGDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTADHD 125
LL F K+ IG G K + SP I + KQP VL ++ D
Sbjct 194 VLLPFLKWFIG-------GSGTKSLKLLNFVVRSPWKTI--DAIAEIKQP-VLFLSGLQD 243
Query 126 DRVSPFHSLKYIAELQHAAGYSSPVSIPLIAHIDTKAGHG 165
+ V PFH A+ + V P H+DT G
Sbjct 244 EMVPPFHMKMLYAKAAARNPQCTFVEFPSGMHMDTWLSGG 283
> tgo:TGME49_046800 acylamino-acid-releasing enzyme, putative
(EC:1.7.2.2 3.4.19.1); K01303 acylaminoacyl-peptidase [EC:3.4.19.1]
Length=851
Score = 33.9 bits (76), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 29/99 (29%), Positives = 41/99 (41%), Gaps = 13/99 (13%)
Query 28 SPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLLRFHKFTIGHAWVSDYGDPD 87
+P + ++GGS+GG L Q PDLF A + V +L + W + G
Sbjct 681 TPARTVVVGGSHGGFLTCHLIGQFPDLFAAASTRNPVTNLASMVVESDIPDWCAAEGLHR 740
Query 88 K--------EEDFTHIYKISPLHNIKPEVAQSDKQPAVL 118
K E D +YK SP+ AQ K P +L
Sbjct 741 KFHPSFGLTENDIVALYKASPV-----AYAQHVKTPLLL 774
> dre:100334632 dachsous 1-like
Length=3237
Score = 31.6 bits (70), Expect = 1.7, Method: Composition-based stats.
Identities = 28/91 (30%), Positives = 42/91 (46%), Gaps = 8/91 (8%)
Query 76 GHAWVSDYGDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTADHDDRVSPFHSLK 135
G WV D D EE T ++ +K E A S AV ++ D +D + F
Sbjct 2074 GEIWVEDSTGLDFEE--TPRLRLV----VKAETASSSSFMAVNLLLQDVNDNLPRFQLHN 2127
Query 136 YIAELQHAAGYSSPVSIPLIAHIDTKAGHGG 166
Y+A ++ A GY P+ I ++A+ D G G
Sbjct 2128 YVAYVREAQGYDHPI-IQVLAN-DLDQGQNG 2156
> mmu:74129 Dmgdh, 1200014D15Rik, AI787269, MGC107623; dimethylglycine
dehydrogenase precursor (EC:1.5.99.2); K00315 dimethylglycine
dehydrogenase [EC:1.5.99.2]
Length=869
Score = 31.2 bits (69), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 24/98 (24%), Positives = 44/98 (44%), Gaps = 9/98 (9%)
Query 55 FRAVVAKVGVMDLLRFHKFTIGHAWVSD-----YGDPDKEEDFTHI-YKISPLHNIKPEV 108
++ V+ +VGV+DL F KF I + + + + FT+I + ++P + E+
Sbjct 515 YKQVMQRVGVIDLSPFGKFNIKGRDSTQLLDHLFANVIPKVGFTNISHMLTPRGRVYAEL 574
Query 109 AQSDKQPAVLVMTADHDDRVSPFHSLKYIAELQHAAGY 146
S + P ++ S H L++I E GY
Sbjct 575 TVSQQSPGEFLLITGSG---SELHDLRWIEEAAFRGGY 609
> mmu:74388 Dpp8, 2310004I03Rik, 4932434F09Rik, AI666706, DPP_VIII;
dipeptidylpeptidase 8 (EC:3.4.14.5); K08655 dipeptidyl-peptidase
8 [EC:3.4.14.5]
Length=892
Score = 30.4 bits (67), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 26/54 (48%), Gaps = 1/54 (1%)
Query 8 QVSYDDFQSAAEYLFKK-GYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVA 60
Q+ DD +YL + + D++ I G S GG L QR D+FR +A
Sbjct 718 QIEIDDQVEGLQYLASQYDFIDLDRVGIHGWSYGGYLSLMALMQRSDIFRVAIA 771
> xla:432175 dpp10, MGC84485; dipeptidyl-peptidase 10 (non-functional)
Length=796
Score = 30.4 bits (67), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 34/137 (24%), Positives = 56/137 (40%), Gaps = 13/137 (9%)
Query 9 VSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLL 68
V D +A E+L K+ + P++L+I G GG + LF+ + D +
Sbjct 621 VEVKDQIAAVEWLLKEPFIDPNRLSIFGKGYGGYIASMILKANDRLFKCGALFAPITD-M 679
Query 69 RFHKFTIGHAWVSDYGDPDKEEDFTHIYKISP-LHNIKPEVAQSDKQPAVLVMTADHDDR 127
R + ++ G P +EE Y+ S LHN+ K +L++ D +
Sbjct 680 RLYASAFSERYL---GLPSREEI---TYQASSVLHNV-----HVLKDKNLLLIHGTADAK 728
Query 128 VSPFHSLKYIAELQHAA 144
V HS + I L A
Sbjct 729 VHFQHSAELIKHLVKAG 745
> hsa:57628 DPP10, DPL2, DPPY, DPRP3; dipeptidyl-peptidase 10
(non-functional)
Length=789
Score = 30.4 bits (67), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 35/136 (25%), Positives = 52/136 (38%), Gaps = 13/136 (9%)
Query 9 VSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLL 68
V D +A ++L K Y +L+I G GG + LF+ + DL
Sbjct 615 VEVKDQITAVKFLLKLPYIDSKRLSIFGKGYGGYIASMILKSDEKLFKCGSVVAPITDLK 674
Query 69 RFHKFTIGHAWVSDY-GDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTADHDDR 127
+ A+ Y G P KEE S LHN+ K+ +L++ D +
Sbjct 675 LY-----ASAFSERYLGMPSKEESTYQ--AASVLHNV-----HGLKEENILIIHGTADTK 722
Query 128 VSPFHSLKYIAELQHA 143
V HS + I L A
Sbjct 723 VHFQHSAELIKHLIKA 738
> mmu:74032 Sdr42e1, 4632417N05Rik, AI117603; short chain dehydrogenase/reductase
family 42E, member 1 (EC:1.1.1.-)
Length=394
Score = 29.6 bits (65), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 21/79 (26%), Positives = 35/79 (44%), Gaps = 14/79 (17%)
Query 83 YGDPDKEEDFTHIYKISPLHNIKPEVAQSDK------QPAVLVMTADHDDRVSPFHSLKY 136
YGDP +F H+ ++ H + E ++DK QP + D R P ++ ++
Sbjct 214 YGDPQSLVEFVHVDNLAKAHILASEALKADKGHVASGQPYFI-----SDGR--PVNNFEF 266
Query 137 IAELQHAAGYSSP-VSIPL 154
L GY+ P +PL
Sbjct 267 FRPLVEGLGYTFPSTRLPL 285
> dre:322670 aco2, cb1017, wu:fa10e03, wu:fb69g04, wu:fc20c11;
aconitase 2, mitochondrial (EC:4.2.1.3); K01681 aconitate hydratase
1 [EC:4.2.1.3]
Length=782
Score = 29.6 bits (65), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 30/63 (47%), Gaps = 2/63 (3%)
Query 126 DRVSPFHSLKYIAELQHAAGYSSPVSIPLIAHID--TKAGHGGGKPLMKVLEEQADTYGF 183
DRV+ + +A LQ + V++P H D +A GG + L K E + Y F
Sbjct 96 DRVAMQDATAQMAMLQFISSGLPKVAVPSTIHCDHLIEAQTGGAQDLQKAKEVNQEVYSF 155
Query 184 IAS 186
+AS
Sbjct 156 LAS 158
Lambda K H
0.317 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6060209704
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40