bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0610_orf2
Length=136
Score E
Sequences producing significant alignments: (Bits) Value
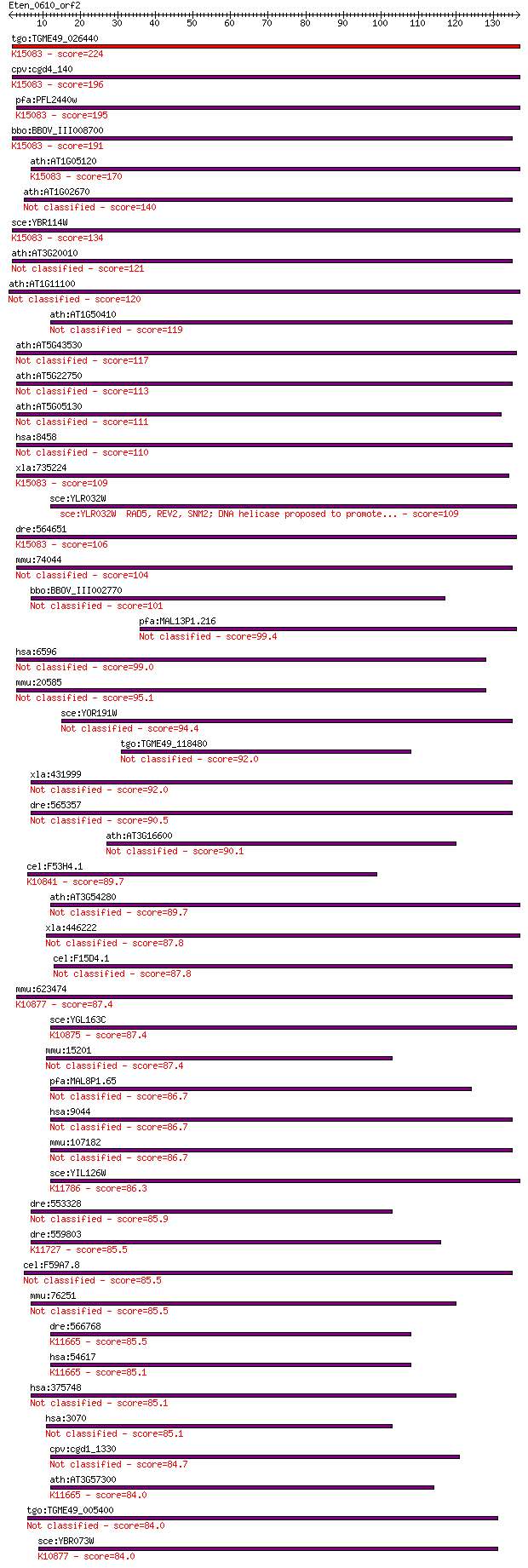
tgo:TGME49_026440 SNF2/RAD54 helicase family protein (EC:2.7.1... 224 8e-59
cpv:cgd4_140 Swi2/Snf2 ATpase,Rad16 ortholog ; K15083 DNA repa... 196 2e-50
pfa:PFL2440w DNA repair protein rhp16, putative; K15083 DNA re... 195 3e-50
bbo:BBOV_III008700 17.m10681; DNA repair protein rhp16; K15083... 191 4e-49
ath:AT1G05120 SNF2 domain-containing protein / helicase domain... 170 1e-42
ath:AT1G02670 DNA repair protein, putative 140 8e-34
sce:YBR114W RAD16, PSO5; Protein that recognizes and binds dam... 134 8e-32
ath:AT3G20010 SNF2 domain-containing protein / helicase domain... 121 7e-28
ath:AT1G11100 SNF2 domain-containing protein / helicase domain... 120 1e-27
ath:AT1G50410 SNF2 domain-containing protein / helicase domain... 119 3e-27
ath:AT5G43530 SNF2 domain-containing protein / helicase domain... 117 1e-26
ath:AT5G22750 RAD5; RAD5; ATP binding / ATP-dependent helicase... 113 2e-25
ath:AT5G05130 SNF2 domain-containing protein / helicase domain... 111 7e-25
hsa:8458 TTF2, HuF2; transcription termination factor, RNA pol... 110 9e-25
xla:735224 hltf, MGC131155, smarca3; helicase-like transcripti... 109 3e-24
sce:YLR032W RAD5, REV2, SNM2; DNA helicase proposed to promote... 109 3e-24
dre:564651 hltf; helicase-like transcription factor-like; K150... 106 2e-23
mmu:74044 Ttf2, 4632434F22Rik, 8030447N19, AV218430; transcrip... 104 8e-23
bbo:BBOV_III002770 17.m07263; SNF2 family N-terminal domain co... 101 7e-22
pfa:MAL13P1.216 DNA helicase, putative 99.4 3e-21
hsa:6596 HLTF, HIP116, HIP116A, HLTF1, RNF80, SMARCA3, SNF2L3,... 99.0 4e-21
mmu:20585 Hltf, AF010600, BC057116, P113, Smarca3, Snf2l3; hel... 95.1 6e-20
sce:YOR191W ULS1, DIS1, RIS1, TID4; RING finger protein involv... 94.4 8e-20
tgo:TGME49_118480 DNA repair protein, putative (EC:2.7.11.1) 92.0 5e-19
xla:431999 ttf2, MGC81081; transcription termination factor, R... 92.0 5e-19
dre:565357 btaf1, TAFII, wu:fd60h10, wu:fe26c12; BTAF1 RNA pol... 90.5 1e-18
ath:AT3G16600 SNF2 domain-containing protein / helicase domain... 90.1 2e-18
cel:F53H4.1 csb-1; human CSB (Cockayne Syndrome B) homolog fam... 89.7 2e-18
ath:AT3G54280 RGD3; RGD3 (ROOT GROWTH DEFECTIVE 3); ATP bindin... 89.7 3e-18
xla:446222 hells, lsh, nbla10143, pasg, smarca6; helicase, lym... 87.8 9e-18
cel:F15D4.1 btf-1; BTAF (TBP-associated factor) homolog family... 87.8 1e-17
mmu:623474 Rad54b, E130016E03Rik, Fsbp, MGC67261; RAD54 homolo... 87.4 1e-17
sce:YGL163C RAD54, XRS1; DNA-dependent ATPase, stimulates stra... 87.4 1e-17
mmu:15201 Hells, AI323785, E130115I21Rik, LSH, Lysh, PASG, YFK... 87.4 1e-17
pfa:MAL8P1.65 DNA helicase, putative 86.7 2e-17
hsa:9044 BTAF1, KIAA0940, MGC138406, MOT1, TAF(II)170, TAF172,... 86.7 2e-17
mmu:107182 Btaf1, AI414500, AI447930, E430027O22Rik, TAF170; B... 86.7 2e-17
sce:YIL126W STH1, NPS1; ATPase component of the RSC chromatin ... 86.3 3e-17
dre:553328 hells, cb65, im:6911667, pasg, sb:cb65, sb:cb749; h... 85.9 4e-17
dre:559803 novel protein similar to SWI/SNF related, matrix as... 85.5 4e-17
cel:F59A7.8 hypothetical protein 85.5 4e-17
mmu:76251 0610007P08Rik, 1700019D06Rik, 9330134C04Rik, MGC1001... 85.5 4e-17
dre:566768 si:ch211-244p18.3; K11665 DNA helicase INO80 [EC:3.... 85.5 5e-17
hsa:54617 INO80, INO80A, INOC1, hINO80; INO80 homolog (S. cere... 85.1 5e-17
hsa:375748 C9orf102, FLJ37706, MGC30192, MGC43364, RAD26L, SR2... 85.1 6e-17
hsa:3070 HELLS, FLJ10339, LSH, PASG, SMARCA6; helicase, lympho... 85.1 6e-17
cpv:cgd1_1330 SWI/SNF-related, matrix associated, actin-depend... 84.7 8e-17
ath:AT3G57300 INO80; INO80 (INO80 ORTHOLOG); ATP binding / DNA... 84.0 1e-16
tgo:TGME49_005400 DNA repair protein RAD54, putative (EC:2.7.1... 84.0 1e-16
sce:YBR073W RDH54, TID1; DNA-dependent ATPase, stimulates stra... 84.0 1e-16
> tgo:TGME49_026440 SNF2/RAD54 helicase family protein (EC:2.7.11.1
6.1.1.11); K15083 DNA repair protein RAD16
Length=1667
Score = 224 bits (570), Expect = 8e-59, Method: Compositional matrix adjust.
Identities = 106/136 (77%), Positives = 121/136 (88%), Gaps = 1/136 (0%)
Query 2 RVKREGIHCAQLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMD 61
R+K+ GIHCA++ GSM+ V+RSNVLYAFNND +LKV+LISLKAGGEGLNLQ+ASRI+LMD
Sbjct 1532 RLKKGGIHCAKMVGSMSIVSRSNVLYAFNNDPSLKVLLISLKAGGEGLNLQIASRIFLMD 1591
Query 62 PWWNPAAEMQAIQRAHRIGQT-RPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQ 120
PWWNPAAEMQAIQRAHRIGQ + V +RFI TIEERILQLQEKKQLVFDGTVGA +
Sbjct 1592 PWWNPAAEMQAIQRAHRIGQRHKEVIAIRFIAEKTIEERILQLQEKKQLVFDGTVGACDH 1651
Query 121 AMHKLTQEDLRFLFQS 136
AM KLTQ+DLRFLFQ+
Sbjct 1652 AMTKLTQDDLRFLFQN 1667
> cpv:cgd4_140 Swi2/Snf2 ATpase,Rad16 ortholog ; K15083 DNA repair
protein RAD16
Length=1278
Score = 196 bits (498), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 88/135 (65%), Positives = 111/135 (82%), Gaps = 0/135 (0%)
Query 2 RVKREGIHCAQLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMD 61
R+K+ I C L GSM+ + R+++LY+FN +LK+ILISLKAGGEGLNLQVA+ ++L+D
Sbjct 1144 RLKKANIGCVMLAGSMSILQRNSILYSFNKFPDLKIILISLKAGGEGLNLQVANYVFLLD 1203
Query 62 PWWNPAAEMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQA 121
PWWNPA E+QA QRAHRIGQ + V +RFI DTIEER+ QLQEKKQLVFDGTVGASN A
Sbjct 1204 PWWNPAVELQAFQRAHRIGQKKKVTALRFITKDTIEERMFQLQEKKQLVFDGTVGASNNA 1263
Query 122 MHKLTQEDLRFLFQS 136
++KL +DL+FLFQ+
Sbjct 1264 LNKLNSDDLKFLFQN 1278
> pfa:PFL2440w DNA repair protein rhp16, putative; K15083 DNA
repair protein RAD16
Length=1647
Score = 195 bits (496), Expect = 3e-50, Method: Composition-based stats.
Identities = 86/134 (64%), Positives = 111/134 (82%), Gaps = 0/134 (0%)
Query 3 VKREGIHCAQLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDP 62
+K+ I C++L G M+ ++R+N+LY FN D L+V+LISLKAGGEGLNLQVA+RI+++DP
Sbjct 1514 LKKHNIVCSKLLGYMSMISRNNILYNFNQDKQLRVLLISLKAGGEGLNLQVANRIFIVDP 1573
Query 63 WWNPAAEMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQAM 122
WWNPAAE+QAIQRAHRIGQT+ V +RFI +T+EE+I+QLQ KKQLVFD T+G S AM
Sbjct 1574 WWNPAAELQAIQRAHRIGQTKTVYAIRFIIENTVEEKIIQLQNKKQLVFDSTIGDSGNAM 1633
Query 123 HKLTQEDLRFLFQS 136
KL++EDL FLF S
Sbjct 1634 QKLSKEDLAFLFHS 1647
> bbo:BBOV_III008700 17.m10681; DNA repair protein rhp16; K15083
DNA repair protein RAD16
Length=1289
Score = 191 bits (486), Expect = 4e-49, Method: Compositional matrix adjust.
Identities = 85/133 (63%), Positives = 106/133 (79%), Gaps = 0/133 (0%)
Query 2 RVKREGIHCAQLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMD 61
R+K I CA L G+ +R N+L FN + +L+V+LISL AGGEGLNLQ+A+RI+LMD
Sbjct 1155 RLKTANIECAVLVGNTKIESRRNILLEFNKNPSLRVMLISLNAGGEGLNLQIANRIFLMD 1214
Query 62 PWWNPAAEMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQA 121
PWWNPAAE+QAIQRAHRIGQT+PV +RFIC DTIEERI+ LQEKK ++FD T+ +S ++
Sbjct 1215 PWWNPAAELQAIQRAHRIGQTKPVYAIRFICKDTIEERIIALQEKKMILFDATICSSGES 1274
Query 122 MHKLTQEDLRFLF 134
M KLT EDL FLF
Sbjct 1275 MKKLTSEDLSFLF 1287
> ath:AT1G05120 SNF2 domain-containing protein / helicase domain-containing
protein / RING finger domain-containing protein;
K15083 DNA repair protein RAD16
Length=833
Score = 170 bits (431), Expect = 1e-42, Method: Compositional matrix adjust.
Identities = 76/130 (58%), Positives = 98/130 (75%), Gaps = 0/130 (0%)
Query 7 GIHCAQLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDPWWNP 66
G+ C QL GSMT AR + F D + +V L+SLKAGG LNL VAS +++MDPWWNP
Sbjct 704 GVSCVQLVGSMTMAARDTAINKFKEDPDCRVFLMSLKAGGVALNLTVASHVFMMDPWWNP 763
Query 67 AAEMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQAMHKLT 126
A E QA R HRIGQ +P++ VRFI +T+EERIL+LQ+KK+LVF+GTVG S +A+ KLT
Sbjct 764 AVERQAQDRIHRIGQYKPIRVVRFIIENTVEERILRLQKKKELVFEGTVGGSQEAIGKLT 823
Query 127 QEDLRFLFQS 136
+ED+RFLF +
Sbjct 824 EEDMRFLFTT 833
> ath:AT1G02670 DNA repair protein, putative
Length=678
Score = 140 bits (354), Expect = 8e-34, Method: Compositional matrix adjust.
Identities = 64/131 (48%), Positives = 92/131 (70%), Gaps = 1/131 (0%)
Query 5 REGIHCAQLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDPWW 64
+ G+ C QL GSM+ A+ L F + + +V+L+SL+AGG LNL AS +++MDPWW
Sbjct 547 KSGVSCVQLVGSMSKAAKDAALKNFKEEPDCRVLLMSLQAGGVALNLTAASHVFMMDPWW 606
Query 65 NPAAEMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQA-MH 123
NPA E QA R HRIGQ +PV+ VRFI T+EE+IL LQ+KK+ +F+ T+G S +A +
Sbjct 607 NPAVERQAQDRIHRIGQCKPVRVVRFIMEKTVEEKILTLQKKKEDLFESTLGDSEEAVVQ 666
Query 124 KLTQEDLRFLF 134
KL ++D++ LF
Sbjct 667 KLGEDDIKSLF 677
> sce:YBR114W RAD16, PSO5; Protein that recognizes and binds damaged
DNA in an ATP-dependent manner (with Rad7p) during nucleotide
excision repair; subunit of Nucleotide Excision Repair
Factor 4 (NEF4) and the Elongin-Cullin-Socs (ECS) ligase
complex (EC:3.6.1.-); K15083 DNA repair protein RAD16
Length=790
Score = 134 bits (337), Expect = 8e-32, Method: Compositional matrix adjust.
Identities = 63/135 (46%), Positives = 86/135 (63%), Gaps = 0/135 (0%)
Query 2 RVKREGIHCAQLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMD 61
R+KR G +L GSM+ R + F N+ +V L+SLKAGG LNL AS+++++D
Sbjct 656 RLKRAGFQTVKLQGSMSPTQRDETIKYFMNNIQCEVFLVSLKAGGVALNLCEASQVFILD 715
Query 62 PWWNPAAEMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQA 121
PWWNP+ E Q+ R HRIGQ RPVK RF D+IE RI++LQEKK + T+ A
Sbjct 716 PWWNPSVEWQSGDRVHRIGQYRPVKITRFCIEDSIEARIIELQEKKANMIHATINQDEAA 775
Query 122 MHKLTQEDLRFLFQS 136
+ +LT DL+FLF +
Sbjct 776 ISRLTPADLQFLFNN 790
> ath:AT3G20010 SNF2 domain-containing protein / helicase domain-containing
protein / RING finger domain-containing protein
Length=1047
Score = 121 bits (303), Expect = 7e-28, Method: Compositional matrix adjust.
Identities = 60/135 (44%), Positives = 85/135 (62%), Gaps = 2/135 (1%)
Query 2 RVKREGIHCAQLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMD 61
R+ GI +L G+M+ AR + F+ ++KV+L+SLKAG GLN+ A + L+D
Sbjct 911 RILESGIEFRRLDGTMSLAARDRAVKEFSKKPDVKVMLMSLKAGNLGLNMVAACHVILLD 970
Query 62 PWWNPAAEMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASN-- 119
WWNP E QAI RAHRIGQTRPV R DT+E+RIL+LQE+K+ + G +
Sbjct 971 LWWNPTTEDQAIDRAHRIGQTRPVTVTRITIKDTVEDRILKLQEEKRTMVASAFGEEHGG 1030
Query 120 QAMHKLTQEDLRFLF 134
+ +LT +DL++LF
Sbjct 1031 SSATRLTVDDLKYLF 1045
> ath:AT1G11100 SNF2 domain-containing protein / helicase domain-containing
protein / zinc finger protein-related
Length=1226
Score = 120 bits (301), Expect = 1e-27, Method: Composition-based stats.
Identities = 61/138 (44%), Positives = 82/138 (59%), Gaps = 2/138 (1%)
Query 1 ARVKREGIHCAQLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLM 60
A +K GI + G MT AR + FN ++ V+++SLKA GLN+ A + ++
Sbjct 1089 AGLKSSGIQYRRFDGKMTVPARDAAVQDFNTLPDVSVMIMSLKAASLGLNMVAACHVIML 1148
Query 61 DPWWNPAAEMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQ 120
D WWNP E QAI RAHRIGQTRPVK VRF DT+E+RIL LQ+KK+ + G
Sbjct 1149 DLWWNPTTEDQAIDRAHRIGQTRPVKVVRFTVKDTVEDRILALQQKKRKMVASAFGEHEN 1208
Query 121 AMHK--LTQEDLRFLFQS 136
+ L+ EDL +LF +
Sbjct 1209 GSRESHLSVEDLNYLFMA 1226
> ath:AT1G50410 SNF2 domain-containing protein / helicase domain-containing
protein / RING finger domain-containing protein
Length=981
Score = 119 bits (297), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 57/125 (45%), Positives = 83/125 (66%), Gaps = 2/125 (1%)
Query 12 QLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDPWWNPAAEMQ 71
+L G+M+ +AR + F+ND ++KV+++SLKAG GLN+ A + L+D WWNP E Q
Sbjct 855 RLDGTMSLIARDRAVKEFSNDPDVKVMIMSLKAGNLGLNMIAACHVILLDLWWNPTTEDQ 914
Query 72 AIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASN--QAMHKLTQED 129
AI RAHRIGQTRPV R +T+E+RIL LQE+K+ + G + + +LT +D
Sbjct 915 AIDRAHRIGQTRPVTVTRITIKNTVEDRILALQEEKRKMVASAFGEDHGGSSATRLTVDD 974
Query 130 LRFLF 134
L++LF
Sbjct 975 LKYLF 979
> ath:AT5G43530 SNF2 domain-containing protein / helicase domain-containing
protein / RING finger domain-containing protein
Length=1277
Score = 117 bits (292), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 59/133 (44%), Positives = 81/133 (60%), Gaps = 2/133 (1%)
Query 3 VKREGIHCAQLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDP 62
++R G + G + R VL FN ++L+SLKAGG GLNL AS ++LMDP
Sbjct 1147 LRRRGFEFLRFDGKLAQKGREKVLKEFNETKQKTILLMSLKAGGVGLNLTAASSVFLMDP 1206
Query 63 WWNPAAEMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQAM 122
WWNPA E QAI R HRIGQ R V RFI DT+EER+ Q+Q +KQ + G + +++ +
Sbjct 1207 WWNPAVEEQAIMRIHRIGQKRTVFVRRFIVKDTVEERMQQVQARKQRMIAGAL--TDEEV 1264
Query 123 HKLTQEDLRFLFQ 135
E+L+ LF+
Sbjct 1265 RSARLEELKMLFR 1277
> ath:AT5G22750 RAD5; RAD5; ATP binding / ATP-dependent helicase/
DNA binding / helicase/ hydrolase, acting on acid anhydrides,
in phosphorus-containing anhydrides / nucleic acid binding
/ protein binding / zinc ion binding
Length=1029
Score = 113 bits (283), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 57/132 (43%), Positives = 83/132 (62%), Gaps = 2/132 (1%)
Query 3 VKREGIHCAQLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDP 62
+ R +L G+++ R VL F+ D ++ V+L+SLKAGG G+NL AS ++MDP
Sbjct 899 LSRNNFSFVRLDGTLSQQQREKVLKEFSEDGSILVLLMSLKAGGVGINLTAASNAFVMDP 958
Query 63 WWNPAAEMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQAM 122
WWNPA E QA+ R HRIGQT+ VK RFI T+EER+ +Q +KQ + G + ++Q +
Sbjct 959 WWNPAVEEQAVMRIHRIGQTKEVKIRRFIVKGTVEERMEAVQARKQRMISGAL--TDQEV 1016
Query 123 HKLTQEDLRFLF 134
E+L+ LF
Sbjct 1017 RSARIEELKMLF 1028
> ath:AT5G05130 SNF2 domain-containing protein / helicase domain-containing
protein / RING finger domain-containing protein
Length=862
Score = 111 bits (277), Expect = 7e-25, Method: Compositional matrix adjust.
Identities = 59/131 (45%), Positives = 81/131 (61%), Gaps = 9/131 (6%)
Query 3 VKREGIHCAQLTGSMTAVARSNVLYAFNND--TNLKVILISLKAGGEGLNLQVASRIYLM 60
+K G +L G+MT R+ V+ F N T V+L SLKA G G+NL ASR+YL
Sbjct 726 LKAAGFTILRLDGAMTVKKRTQVIGEFGNPELTGPVVLLASLKASGTGINLTAASRVYLF 785
Query 61 DPWWNPAAEMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQ 120
DPWWNPA E QA+ R HRIGQ + VK +R I ++IEER+L+LQ+KK+ +N+
Sbjct 786 DPWWNPAVEEQAMDRIHRIGQKQEVKMIRMIARNSIEERVLELQQKKK-------NLANE 838
Query 121 AMHKLTQEDLR 131
A + ++D R
Sbjct 839 AFKRRQKKDER 849
> hsa:8458 TTF2, HuF2; transcription termination factor, RNA polymerase
II
Length=1162
Score = 110 bits (276), Expect = 9e-25, Method: Compositional matrix adjust.
Identities = 57/132 (43%), Positives = 85/132 (64%), Gaps = 0/132 (0%)
Query 3 VKREGIHCAQLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDP 62
+K+ G+ A + GS+ R +++ AFN+ +V+LISL AGG GLNL + ++L+D
Sbjct 1029 LKKHGLTYATIDGSVNPKQRMDLVEAFNHSRGPQVMLISLLAGGVGLNLTGGNHLFLLDM 1088
Query 63 WWNPAAEMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQAM 122
WNP+ E QA R +R+GQ + V RF+C T+EE+ILQLQEKK+ + + S +++
Sbjct 1089 HWNPSLEDQACDRIYRVGQQKDVVIHRFVCEGTVEEKILQLQEKKKDLAKQVLSGSGESV 1148
Query 123 HKLTQEDLRFLF 134
KLT DLR LF
Sbjct 1149 TKLTLADLRVLF 1160
> xla:735224 hltf, MGC131155, smarca3; helicase-like transcription
factor; K15083 DNA repair protein RAD16
Length=999
Score = 109 bits (272), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 56/136 (41%), Positives = 84/136 (61%), Gaps = 5/136 (3%)
Query 3 VKREGIHCAQLTGSMTAVARSNVLYAFN--NDTNLKVILISLKAGGEGLNLQVASRIYLM 60
++ G +L GSMT R+ + +F + + ++L+SLKAGG GLNL ASR++LM
Sbjct 861 LRESGFMFTRLDGSMTQKKRTEAIQSFQRPDAQSPTIMLLSLKAGGVGLNLTAASRVFLM 920
Query 61 DPWWNPAAEMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQ 120
DP WNPAAE Q R HR+GQT+ V +F+ D++EE +L++Q KK+ + G GA
Sbjct 921 DPAWNPAAEEQCFDRCHRLGQTKEVIITKFVVRDSVEENMLKIQSKKRQLAAGAFGAKKS 980
Query 121 AMHKLTQ---EDLRFL 133
+ ++ Q ED+R L
Sbjct 981 SASQIKQARIEDIRTL 996
> sce:YLR032W RAD5, REV2, SNM2; DNA helicase proposed to promote
replication fork regression during postreplication repair
by template switching; RING finger containing ubiquitin ligase;
stimulates the synthesis of free and PCNA-bound polyubiquitin
chains by Ubc13p-Mms2p (EC:3.6.1.-); K01529 [EC:3.6.1.-]
Length=1169
Score = 109 bits (272), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 55/126 (43%), Positives = 83/126 (65%), Gaps = 3/126 (2%)
Query 12 QLTGSMTAVARSNVL--YAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDPWWNPAAE 69
+ G ++ R++VL +A + + K++L+SLKAGG GLNL AS Y+MDPWW+P+ E
Sbjct 1045 KFDGRLSLKERTSVLADFAVKDYSRQKILLLSLKAGGVGLNLTCASHAYMMDPWWSPSME 1104
Query 70 MQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQAMHKLTQED 129
QAI R HRIGQT VK +RFI D+IEE++L++QEKK+ + + + K E+
Sbjct 1105 DQAIDRLHRIGQTNSVKVMRFIIQDSIEEKMLRIQEKKRTIGEA-MDTDEDERRKRRIEE 1163
Query 130 LRFLFQ 135
++ LF+
Sbjct 1164 IQMLFE 1169
> dre:564651 hltf; helicase-like transcription factor-like; K15083
DNA repair protein RAD16
Length=942
Score = 106 bits (264), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 54/136 (39%), Positives = 85/136 (62%), Gaps = 3/136 (2%)
Query 3 VKREGIHCAQLTGSMTAVARSNVLYAFNNDT--NLKVILISLKAGGEGLNLQVASRIYLM 60
++ G +L GS+ AR+ + F + T + ++L+SLKAGG GLNL AS++++M
Sbjct 806 LREYGFSFTRLDGSLIQRARAKAIEDFQDSTPGSPTIMLLSLKAGGVGLNLTAASQVFVM 865
Query 61 DPWWNPAAEMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQ 120
DP WNPAAE Q + R HR+GQ+R V +FI D++EE ++++Q+KKQ + D G N
Sbjct 866 DPAWNPAAEDQCVDRCHRLGQSRDVVITKFIVKDSVEENMVKIQKKKQELVDKAFGVKNS 925
Query 121 AMHKLTQ-EDLRFLFQ 135
K + +D+R L +
Sbjct 926 QDAKQARIDDIRALME 941
> mmu:74044 Ttf2, 4632434F22Rik, 8030447N19, AV218430; transcription
termination factor, RNA polymerase II
Length=1138
Score = 104 bits (259), Expect = 8e-23, Method: Compositional matrix adjust.
Identities = 55/132 (41%), Positives = 82/132 (62%), Gaps = 0/132 (0%)
Query 3 VKREGIHCAQLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDP 62
+K+ + A + GS+ R +++ AFN+ +V+LISL AGG GLNL + ++L+D
Sbjct 1005 LKKNRLTYATIDGSVNPKQRMDLVEAFNHSQGPQVMLISLLAGGVGLNLTGGNHLFLLDM 1064
Query 63 WWNPAAEMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQAM 122
WNP+ E QA R +R+GQ + V RF+C T+EE+ILQLQEKK+ + + S +
Sbjct 1065 HWNPSLEDQACDRIYRVGQKKDVVIHRFVCEGTVEEKILQLQEKKKDLAKQVLSGSEGPV 1124
Query 123 HKLTQEDLRFLF 134
KLT DL+ LF
Sbjct 1125 TKLTLADLKILF 1136
> bbo:BBOV_III002770 17.m07263; SNF2 family N-terminal domain
containing protein
Length=860
Score = 101 bits (251), Expect = 7e-22, Method: Compositional matrix adjust.
Identities = 52/115 (45%), Positives = 69/115 (60%), Gaps = 5/115 (4%)
Query 7 GIHCAQL-----TGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMD 61
G+H QL G R++++ F ND N+ ++LIS KAGG GLNL VAS + LMD
Sbjct 736 GLHKPQLEYLRLDGGHNPSTRTDIVERFTNDPNITLLLISTKAGGTGLNLTVASTVILMD 795
Query 62 PWWNPAAEMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVG 116
WNP + QA R+HRIGQT PV + +C DTIEE I + ++K L+ D G
Sbjct 796 LDWNPHNDAQAENRSHRIGQTEPVDVYKLMCEDTIEEYIWECCQRKLLLDDAFSG 850
> pfa:MAL13P1.216 DNA helicase, putative
Length=1446
Score = 99.4 bits (246), Expect = 3e-21, Method: Composition-based stats.
Identities = 49/108 (45%), Positives = 65/108 (60%), Gaps = 8/108 (7%)
Query 36 KVILISLKAGGEGLNLQVASRIYLMDPWWNPAAEMQAIQRAHRIGQTRPVKGVRFICTDT 95
KV+L SLKAGG GLNL V+S++YLMD WWNPA E QA +R HRIGQ + V +F+ T
Sbjct 1333 KVLLCSLKAGGVGLNLTVSSKVYLMDLWWNPAIEDQAFERVHRIGQLKEVNIYKFVLEKT 1392
Query 96 IEERILQLQEKKQLVFDGTVGAS--------NQAMHKLTQEDLRFLFQ 135
+EERILQ+ + KQ + + N KL +D +F+
Sbjct 1393 VEERILQIHQSKQYTANQILTQEGNKLNTEMNMVPQKLGMDDFILMFK 1440
> hsa:6596 HLTF, HIP116, HIP116A, HLTF1, RNF80, SMARCA3, SNF2L3,
ZBU1; helicase-like transcription factor
Length=1009
Score = 99.0 bits (245), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 52/127 (40%), Positives = 73/127 (57%), Gaps = 2/127 (1%)
Query 3 VKREGIHCAQLTGSMTAVARSNVLYAFNNDT--NLKVILISLKAGGEGLNLQVASRIYLM 60
+K G +L GSM R + F N + ++L+SLKAGG GLNL ASR++LM
Sbjct 871 LKASGFVFTRLDGSMAQKKRVESIQCFQNTEAGSPTIMLLSLKAGGVGLNLSAASRVFLM 930
Query 61 DPWWNPAAEMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQ 120
DP WNPAAE Q R HR+GQ + V +FI D++EE +L++Q KK+ + G G
Sbjct 931 DPAWNPAAEDQCFDRCHRLGQKQEVIITKFIVKDSVEENMLKIQNKKRELAAGAFGTKKP 990
Query 121 AMHKLTQ 127
++ Q
Sbjct 991 NADEMKQ 997
> mmu:20585 Hltf, AF010600, BC057116, P113, Smarca3, Snf2l3; helicase-like
transcription factor
Length=1003
Score = 95.1 bits (235), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 51/127 (40%), Positives = 72/127 (56%), Gaps = 2/127 (1%)
Query 3 VKREGIHCAQLTGSMTAVARSNVLYAFNNDT--NLKVILISLKAGGEGLNLQVASRIYLM 60
+K G +L GSM R + F N + ++L+SLKAGG GLNL ASR++LM
Sbjct 865 LKASGFVFTRLDGSMAQKKRVESIQRFQNTEAGSPTIMLLSLKAGGVGLNLCAASRVFLM 924
Query 61 DPWWNPAAEMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQ 120
DP WNPAAE Q R HR+GQ + V +FI D++EE +L++Q K+ + G G
Sbjct 925 DPAWNPAAEDQCFDRCHRLGQKQEVIITKFIVKDSVEENMLKIQNTKRDLAAGAFGTKKT 984
Query 121 AMHKLTQ 127
+ + Q
Sbjct 985 DANDMKQ 991
> sce:YOR191W ULS1, DIS1, RIS1, TID4; RING finger protein involved
in proteolytic control of sumoylated substrates; interacts
with SUMO (Smt3p); member of the SWI/SNF family of DNA-dependent
ATPases; plays a role in antagonizing silencing during
mating-type switching (EC:3.6.1.-)
Length=1619
Score = 94.4 bits (233), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 46/121 (38%), Positives = 75/121 (61%), Gaps = 1/121 (0%)
Query 15 GSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDPWWNPAAEMQAIQ 74
GSM A RS+V+ F D +++LIS+KAG GL L A+ + ++DP+WNP E QA
Sbjct 1494 GSMNAQRRSDVINEFYRDPEKRILLISMKAGNSGLTLTCANHVVIVDPFWNPYVEEQAQD 1553
Query 75 RAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASN-QAMHKLTQEDLRFL 133
R +RI QT+ V+ + D++E+RI +LQ++K+ + D + + ++ L + +L FL
Sbjct 1554 RCYRISQTKKVQVHKLFIKDSVEDRISELQKRKKEMVDSAMDPGKIKEVNSLGRRELGFL 1613
Query 134 F 134
F
Sbjct 1614 F 1614
> tgo:TGME49_118480 DNA repair protein, putative (EC:2.7.11.1)
Length=1733
Score = 92.0 bits (227), Expect = 5e-19, Method: Composition-based stats.
Identities = 42/77 (54%), Positives = 55/77 (71%), Gaps = 0/77 (0%)
Query 31 NDTNLKVILISLKAGGEGLNLQVASRIYLMDPWWNPAAEMQAIQRAHRIGQTRPVKGVRF 90
+D +++L SLKAG GLNL ASR YLMD WWNP E QA++R R GQ +PVK VRF
Sbjct 1520 DDEEGRILLCSLKAGNVGLNLTRASRCYLMDGWWNPQVENQAMKRIWRFGQDKPVKVVRF 1579
Query 91 ICTDTIEERILQLQEKK 107
+C T+EER+ +++E K
Sbjct 1580 VCVRTVEERLEEIKEFK 1596
> xla:431999 ttf2, MGC81081; transcription termination factor,
RNA polymerase II
Length=1187
Score = 92.0 bits (227), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 56/129 (43%), Positives = 80/129 (62%), Gaps = 1/129 (0%)
Query 7 GIHCAQLTGSMTAVARSNVLYAFNNDTN-LKVILISLKAGGEGLNLQVASRIYLMDPWWN 65
G+ CA + GS+ R +++ FNN+ +V+L+SL AGG GLNL + ++LMD WN
Sbjct 1057 GLSCATIDGSVNPKQRMDMVEDFNNNPKGPQVMLVSLCAGGVGLNLVGGNHLFLMDMHWN 1116
Query 66 PAAEMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQAMHKL 125
PA E QA R +R+GQ + V RF+C T+EE+I QLQEKK+ + + ++ KL
Sbjct 1117 PALEDQACDRIYRVGQQKDVVIHRFVCLGTVEEKISQLQEKKKDLAKKVLSGNSATFTKL 1176
Query 126 TQEDLRFLF 134
T DLR LF
Sbjct 1177 TLADLRLLF 1185
> dre:565357 btaf1, TAFII, wu:fd60h10, wu:fe26c12; BTAF1 RNA polymerase
II, B-TFIID transcription factor-associated
Length=1858
Score = 90.5 bits (223), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 47/128 (36%), Positives = 74/128 (57%), Gaps = 0/128 (0%)
Query 7 GIHCAQLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDPWWNP 66
G+ +L GS+ A R +++ FNND ++ V+L++ GG GLNL A + ++ WNP
Sbjct 1678 GVTYLRLDGSVQAGLRHSIVSRFNNDPSIDVLLLTTHVGGLGLNLTGADTVVFVEHDWNP 1737
Query 67 AAEMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQAMHKLT 126
++QA+ RAHRIGQ R V R I T+EE+I+ LQ+ K + + + N ++ +
Sbjct 1738 MRDLQAMDRAHRIGQKRVVNVYRLITRGTLEEKIMGLQKFKMTIANTVISQENASLQSMG 1797
Query 127 QEDLRFLF 134
E L LF
Sbjct 1798 TEQLLNLF 1805
> ath:AT3G16600 SNF2 domain-containing protein / helicase domain-containing
protein / RING finger domain-containing protein
Length=638
Score = 90.1 bits (222), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 45/97 (46%), Positives = 60/97 (61%), Gaps = 4/97 (4%)
Query 27 YAFNNDTNLK----VILISLKAGGEGLNLQVASRIYLMDPWWNPAAEMQAIQRAHRIGQT 82
++ N D ++ +L+SLKAG GLN+ AS + L+D WWNP E QAI RAHRIGQT
Sbjct 495 HSTNKDNSISGLVCAMLMSLKAGNLGLNMVAASHVILLDLWWNPTTEDQAIDRAHRIGQT 554
Query 83 RPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASN 119
R V R +T+EERIL L E+K+ + +G N
Sbjct 555 RAVTVTRIAIKNTVEERILTLHERKRNIVASALGEKN 591
> cel:F53H4.1 csb-1; human CSB (Cockayne Syndrome B) homolog family
member (csb-1); K10841 DNA excision repair protein ERCC-6
Length=957
Score = 89.7 bits (221), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 40/93 (43%), Positives = 60/93 (64%), Gaps = 0/93 (0%)
Query 6 EGIHCAQLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDPWWN 65
+GI C LTG+ +A AR ++ F +D ++KV L++ +AGG GLNL A+++ + DP WN
Sbjct 561 KGIKCVSLTGADSAAARPKIIKKFEDDVSIKVFLMTTRAGGLGLNLTCANKVIIFDPDWN 620
Query 66 PAAEMQAIQRAHRIGQTRPVKGVRFICTDTIEE 98
P A+ QA R +R+GQT V R + TIE+
Sbjct 621 PQADNQAKNRIYRMGQTNDVAIYRLVSNGTIED 653
> ath:AT3G54280 RGD3; RGD3 (ROOT GROWTH DEFECTIVE 3); ATP binding
/ DNA binding / binding / helicase/ nucleic acid binding
Length=2038
Score = 89.7 bits (221), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 47/125 (37%), Positives = 71/125 (56%), Gaps = 0/125 (0%)
Query 12 QLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDPWWNPAAEMQ 71
+L GS+ R ++ AFN+D + V+L++ GG GLNL A + M+ WNP + Q
Sbjct 1855 RLDGSVVPEKRFEIVKAFNSDPTIDVLLLTTHVGGLGLNLTSADTLVFMEHDWNPMRDHQ 1914
Query 72 AIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQAMHKLTQEDLR 131
A+ RAHR+GQ R V R I T+EE+++ LQ+ K V + + A N +M + + L
Sbjct 1915 AMDRAHRLGQKRVVNVHRLIMRGTLEEKVMSLQKFKVSVANTVINAENASMKTMNTDQLL 1974
Query 132 FLFQS 136
LF S
Sbjct 1975 DLFAS 1979
> xla:446222 hells, lsh, nbla10143, pasg, smarca6; helicase, lymphoid-specific
Length=838
Score = 87.8 bits (216), Expect = 9e-18, Method: Compositional matrix adjust.
Identities = 48/135 (35%), Positives = 74/135 (54%), Gaps = 9/135 (6%)
Query 11 AQLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDPWWNPAAEM 70
+L GSM+ R + +FN D ++ + L+S +AGG G+NL A + + D WNP A++
Sbjct 646 CRLDGSMSYTDREENMRSFNTDPDVFIFLVSTRAGGLGINLTAADTVIIYDSDWNPQADL 705
Query 71 QAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLV---------FDGTVGASNQA 121
QA R HRIGQTRPV R + +TI+++I++ K+ + F G NQ+
Sbjct 706 QAQDRCHRIGQTRPVVVYRLVTANTIDQKIVERAAAKRKLEKLVIHKNKFKGGQTGINQS 765
Query 122 MHKLTQEDLRFLFQS 136
L ++L L QS
Sbjct 766 KSCLDPQELLELLQS 780
> cel:F15D4.1 btf-1; BTAF (TBP-associated factor) homolog family
member (btf-1)
Length=1649
Score = 87.8 bits (216), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 45/122 (36%), Positives = 68/122 (55%), Gaps = 0/122 (0%)
Query 13 LTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDPWWNPAAEMQA 72
L GS+ A R ++ FN D + V++++ GG GLNL A + +D WNP ++QA
Sbjct 1474 LDGSVPAGDRMKMVNRFNEDKTIDVLILTTHVGGVGLNLTGADTVIFLDHDWNPMKDLQA 1533
Query 73 IQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQAMHKLTQEDLRF 132
I RAHR+GQTR V R I T+EE+++ L + K +GA N +M + +L
Sbjct 1534 IDRAHRLGQTRNVNVYRLITQGTVEEKVMSLAKFKLNTAQALIGADNTSMMTMETGELMN 1593
Query 133 LF 134
+F
Sbjct 1594 MF 1595
> mmu:623474 Rad54b, E130016E03Rik, Fsbp, MGC67261; RAD54 homolog
B (S. cerevisiae); K10877 DNA repair and recombination protein
RAD54B [EC:3.6.4.-]
Length=886
Score = 87.4 bits (215), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 53/134 (39%), Positives = 74/134 (55%), Gaps = 2/134 (1%)
Query 3 VKREGIHCAQLTGSMTAVARSNVLYAFNNDTNLKVI-LISLKAGGEGLNLQVASRIYLMD 61
KR G CA+L G R +++ +FN+ + I L+S KAGG GLNL S + L D
Sbjct 658 CKRHGYACARLDGQTPVSQRQHIVDSFNSKYSTDFIFLLSSKAGGVGLNLIGGSHLILYD 717
Query 62 PWWNPAAEMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQA 121
WNPA ++QA+ R R GQ PV R + T TIEE+I Q Q KQ + V + +
Sbjct 718 IDWNPATDIQAMSRVWRDGQKHPVHIYRLLTTGTIEEKIYQRQISKQGLSGAVVDLTRSS 777
Query 122 MH-KLTQEDLRFLF 134
H + + E+L+ LF
Sbjct 778 EHIQFSVEELKNLF 791
> sce:YGL163C RAD54, XRS1; DNA-dependent ATPase, stimulates strand
exchange by modifying the topology of double-stranded DNA;
involved in the recombinational repair of double-strand
breaks in DNA during vegetative growth and meiosis; member of
the SWI/SNF family (EC:3.6.1.-); K10875 DNA repair and recombination
protein RAD54 and RAD54-like protein [EC:3.6.4.-]
Length=898
Score = 87.4 bits (215), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 52/125 (41%), Positives = 71/125 (56%), Gaps = 1/125 (0%)
Query 12 QLTGSMTAVARSNVLYAFNNDTNLKVI-LISLKAGGEGLNLQVASRIYLMDPWWNPAAEM 70
+L G+M+ R ++ FN+ + I L+S KAGG G+NL A+R+ LMDP WNPAA+
Sbjct 701 RLDGTMSINKRQKLVDRFNDPEGQEFIFLLSSKAGGCGINLIGANRLILMDPDWNPAADQ 760
Query 71 QAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQAMHKLTQEDL 130
QA+ R R GQ + RFI T TIEE+I Q Q K + V A + ++L
Sbjct 761 QALARVWRDGQKKDCFIYRFISTGTIEEKIFQRQSMKMSLSSCVVDAKEDVERLFSSDNL 820
Query 131 RFLFQ 135
R LFQ
Sbjct 821 RQLFQ 825
> mmu:15201 Hells, AI323785, E130115I21Rik, LSH, Lysh, PASG, YFK8;
helicase, lymphoid specific
Length=821
Score = 87.4 bits (215), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 37/92 (40%), Positives = 61/92 (66%), Gaps = 0/92 (0%)
Query 11 AQLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDPWWNPAAEM 70
++L GSM+ R +Y+FN D ++ + L+S +AGG G+NL A + + D WNP +++
Sbjct 626 SRLDGSMSYSEREKNIYSFNTDPDVFLFLVSTRAGGLGINLTAADTVIIYDSDWNPQSDL 685
Query 71 QAIQRAHRIGQTRPVKGVRFICTDTIEERILQ 102
QA R HRIGQT+PV R + +TI+++I++
Sbjct 686 QAQDRCHRIGQTKPVVVYRLVTANTIDQKIVE 717
> pfa:MAL8P1.65 DNA helicase, putative
Length=1221
Score = 86.7 bits (213), Expect = 2e-17, Method: Composition-based stats.
Identities = 46/112 (41%), Positives = 63/112 (56%), Gaps = 1/112 (0%)
Query 12 QLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDPWWNPAAEMQ 71
+L GS + R ++ F+ D N+ V L+S KAGG GLNL A+ + LMD WNP + Q
Sbjct 1100 RLDGSTNTIERQKIIKRFSKDENIFVFLLSTKAGGVGLNLIAANHVILMDQDWNPHNDRQ 1159
Query 72 AIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQAMH 123
A R HR+GQ V R C +TIEE IL+ K +L D G +N+ +
Sbjct 1160 AEDRVHRLGQKNEVFIYRLCCKNTIEEAILKCN-KAKLHLDQAFGGNNELLQ 1210
> hsa:9044 BTAF1, KIAA0940, MGC138406, MOT1, TAF(II)170, TAF172,
TAFII170; BTAF1 RNA polymerase II, B-TFIID transcription
factor-associated, 170kDa (Mot1 homolog, S. cerevisiae)
Length=1849
Score = 86.7 bits (213), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 44/123 (35%), Positives = 71/123 (57%), Gaps = 0/123 (0%)
Query 12 QLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDPWWNPAAEMQ 71
+L GS+ R +++ FNND ++ V+L++ GG GLNL A + ++ WNP ++Q
Sbjct 1676 RLDGSIPPGQRHSIVSRFNNDPSIDVLLLTTHVGGLGLNLTGADTVVFVEHDWNPMRDLQ 1735
Query 72 AIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQAMHKLTQEDLR 131
A+ RAHRIGQ R V R I T+EE+I+ LQ+ K + + + N ++ + + L
Sbjct 1736 AMDRAHRIGQKRVVNVYRLITRGTLEEKIMGLQKFKMNIANTVISQENSSLQSMGTDQLL 1795
Query 132 FLF 134
LF
Sbjct 1796 DLF 1798
> mmu:107182 Btaf1, AI414500, AI447930, E430027O22Rik, TAF170;
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated,
(Mot1 homolog, S. cerevisiae)
Length=1848
Score = 86.7 bits (213), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 44/123 (35%), Positives = 71/123 (57%), Gaps = 0/123 (0%)
Query 12 QLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDPWWNPAAEMQ 71
+L GS+ R +++ FNND ++ V+L++ GG GLNL A + ++ WNP ++Q
Sbjct 1675 RLDGSIPPGQRHSIVSRFNNDPSIDVLLLTTHVGGLGLNLTGADTVVFVEHDWNPMRDLQ 1734
Query 72 AIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQAMHKLTQEDLR 131
A+ RAHRIGQ R V R I T+EE+I+ LQ+ K + + + N ++ + + L
Sbjct 1735 AMDRAHRIGQKRVVNVYRLITRGTLEEKIMGLQKFKMNIANTVISQENSSLQSMGTDQLL 1794
Query 132 FLF 134
LF
Sbjct 1795 DLF 1797
> sce:YIL126W STH1, NPS1; ATPase component of the RSC chromatin
remodeling complex; required for expression of early meiotic
genes; essential helicase-related protein homologous to Snf2p
(EC:3.6.1.-); K11786 ATP-dependent helicase STH1/SNF2 [EC:3.6.4.-]
Length=1359
Score = 86.3 bits (212), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 54/130 (41%), Positives = 76/130 (58%), Gaps = 6/130 (4%)
Query 12 QLTGSMTAVARSNVLYAFN-NDTNLKVILISLKAGGEGLNLQVASRIYLMDPWWNPAAEM 70
+L GS R+ +L AFN D++ L+S +AGG GLNLQ A + + D WNP ++
Sbjct 836 RLDGSTKTEERTEMLNAFNAPDSDYFCFLLSTRAGGLGLNLQTADTVIIFDTDWNPHQDL 895
Query 71 QAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQAMHKLTQED- 129
QA RAHRIGQ V+ +R I TD++EE IL+ +K L DG V + + +K T E+
Sbjct 896 QAQDRAHRIGQKNEVRILRLITTDSVEEVILERAMQK-LDIDGKVIQAGKFDNKSTAEEQ 954
Query 130 ---LRFLFQS 136
LR L +S
Sbjct 955 EAFLRRLIES 964
> dre:553328 hells, cb65, im:6911667, pasg, sb:cb65, sb:cb749;
helicase, lymphoid-specific
Length=853
Score = 85.9 bits (211), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 40/96 (41%), Positives = 60/96 (62%), Gaps = 0/96 (0%)
Query 7 GIHCAQLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDPWWNP 66
G ++L GSM+ R + F++D + + L+S +AGG G+NL A + + D WNP
Sbjct 655 GYEYSRLDGSMSYADRDENMKKFSSDPEVFLFLLSTRAGGLGINLTSADTVIIFDSDWNP 714
Query 67 AAEMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQ 102
A++QA R HRIGQT+PV R I +TI+E+IL+
Sbjct 715 QADLQAQDRCHRIGQTKPVVVHRLITANTIDEKILE 750
> dre:559803 novel protein similar to SWI/SNF related, matrix
associated, actin dependent regulator of chromatin, subfamily
a, member 5 (smarca5); K11727 SWI/SNF-related matrix-associated
actin-dependent regulator of chromatin subfamily A member
1 [EC:3.6.4.-]
Length=1036
Score = 85.5 bits (210), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 47/110 (42%), Positives = 64/110 (58%), Gaps = 2/110 (1%)
Query 7 GIHCAQLTGSMTAVARSNVLYAFNNDTNLKVI-LISLKAGGEGLNLQVASRIYLMDPWWN 65
G +L G+ AR + AFN + K I ++S +AGG G+NL A + L D WN
Sbjct 485 GFEYCRLDGNTPHEAREQAIDAFNAPNSSKFIFMLSTRAGGLGINLATADVVILYDSDWN 544
Query 66 PAAEMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTV 115
P ++QA+ RAHRIGQ +PVK R I +T+EERI++ E K L D V
Sbjct 545 PQVDLQAMDRAHRIGQRKPVKVFRLITDNTVEERIVERAEMK-LRLDSIV 593
> cel:F59A7.8 hypothetical protein
Length=518
Score = 85.5 bits (210), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 53/134 (39%), Positives = 79/134 (58%), Gaps = 5/134 (3%)
Query 5 REGIH-CAQLTGSMTAVARSNVLYAFNNDTN-LKVILISLKAGGEGLNLQVASRIYLMDP 62
+EG H +TG + R + +FN + + +L+SL AGG GLNL + + ++D
Sbjct 376 QEGGHGYTSITGEVAIKDRQERVDSFNQEKGGAQDMLLSLTAGGVGLNLIGGNHLIMVDL 435
Query 63 WWNPAAEMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTV--GASNQ 120
WNPA E QA R +R+GQ + V R I +TIE+R++ LQEKK L +V G++ +
Sbjct 436 HWNPALEQQACDRIYRMGQKKEVHIHRLIVKETIEQRVMSLQEKK-LALAASVLEGSATR 494
Query 121 AMHKLTQEDLRFLF 134
M+KLT D+R LF
Sbjct 495 GMNKLTNSDIRTLF 508
> mmu:76251 0610007P08Rik, 1700019D06Rik, 9330134C04Rik, MGC100170,
Rad26l, Sr278; RIKEN cDNA 0610007P08 gene
Length=699
Score = 85.5 bits (210), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 45/113 (39%), Positives = 66/113 (58%), Gaps = 0/113 (0%)
Query 7 GIHCAQLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDPWWNP 66
G+ +L GS + R ++ FN+ ++ + L+S AGG GLN A+ + L DP WNP
Sbjct 546 GLDYRRLDGSTKSEERLKIVKEFNSSQDVNICLVSTMAGGLGLNFVGANVVILFDPTWNP 605
Query 67 AAEMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASN 119
A ++QA+ RA+RIGQ R VK +R I T+EE + Q KQ + VG+ N
Sbjct 606 ANDLQAVDRAYRIGQCRDVKVLRLISLGTVEEIMYLRQVYKQQLHCVVVGSEN 658
> dre:566768 si:ch211-244p18.3; K11665 DNA helicase INO80 [EC:3.6.4.12]
Length=1552
Score = 85.5 bits (210), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 42/96 (43%), Positives = 59/96 (61%), Gaps = 0/96 (0%)
Query 12 QLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDPWWNPAAEMQ 71
+L GS R +++ F + T++ V L+S +AGG G+NL A + D WNP + Q
Sbjct 1144 RLDGSSKISERRDMVADFQSRTDIFVFLLSTRAGGLGINLTAADTVIFYDSDWNPTVDQQ 1203
Query 72 AIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKK 107
A+ RAHR+GQT+ V R IC TIEERILQ ++K
Sbjct 1204 AMDRAHRLGQTKQVTVYRLICKGTIEERILQRAKEK 1239
> hsa:54617 INO80, INO80A, INOC1, hINO80; INO80 homolog (S. cerevisiae)
(EC:3.6.4.12); K11665 DNA helicase INO80 [EC:3.6.4.12]
Length=1556
Score = 85.1 bits (209), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 42/96 (43%), Positives = 58/96 (60%), Gaps = 0/96 (0%)
Query 12 QLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDPWWNPAAEMQ 71
+L GS R +++ F N ++ V L+S +AGG G+NL A + D WNP + Q
Sbjct 1146 RLDGSSKISERRDMVADFQNRNDIFVFLLSTRAGGLGINLTAADTVIFYDSDWNPTVDQQ 1205
Query 72 AIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKK 107
A+ RAHR+GQT+ V R IC TIEERILQ ++K
Sbjct 1206 AMDRAHRLGQTKQVTVYRLICKGTIEERILQRAKEK 1241
> hsa:375748 C9orf102, FLJ37706, MGC30192, MGC43364, RAD26L, SR278;
chromosome 9 open reading frame 102
Length=712
Score = 85.1 bits (209), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 46/113 (40%), Positives = 66/113 (58%), Gaps = 0/113 (0%)
Query 7 GIHCAQLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDPWWNP 66
G+ +L GS + R ++ FN+ ++ + L+S AGG GLN A+ + L DP WNP
Sbjct 559 GLDYRRLDGSTKSEERLKIVKEFNSTQDVNICLVSTMAGGLGLNFVGANVVVLFDPTWNP 618
Query 67 AAEMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASN 119
A ++QAI RA+RIGQ R VK +R I T+EE + Q KQ + VG+ N
Sbjct 619 ANDLQAIDRAYRIGQCRDVKVLRLISLGTVEEIMYLRQIYKQQLHCVVVGSEN 671
> hsa:3070 HELLS, FLJ10339, LSH, PASG, SMARCA6; helicase, lymphoid-specific
Length=838
Score = 85.1 bits (209), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 36/92 (39%), Positives = 60/92 (65%), Gaps = 0/92 (0%)
Query 11 AQLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDPWWNPAAEM 70
++L GSM+ R +++FN D + + L+S +AGG G+NL A + + D WNP +++
Sbjct 643 SRLDGSMSYSEREKNMHSFNTDPEVFIFLVSTRAGGLGINLTAADTVIIYDSDWNPQSDL 702
Query 71 QAIQRAHRIGQTRPVKGVRFICTDTIEERILQ 102
QA R HRIGQT+PV R + +TI+++I++
Sbjct 703 QAQDRCHRIGQTKPVVVYRLVTANTIDQKIVE 734
> cpv:cgd1_1330 SWI/SNF-related, matrix associated, actin-dependent
regulator of chromatin subfamily A containing DEAD/H box
1
Length=807
Score = 84.7 bits (208), Expect = 8e-17, Method: Composition-based stats.
Identities = 46/109 (42%), Positives = 63/109 (57%), Gaps = 2/109 (1%)
Query 12 QLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDPWWNPAAEMQ 71
+L G+ + R N++ F T + + L+S KA G+GLNL VAS + +MD +NP E Q
Sbjct 698 RLDGTTPILERQNMIEKFQT-TQVPLFLLSTKAAGQGLNLTVASSVIMMDLDYNPQIEKQ 756
Query 72 AIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQ 120
A R HRIGQ++ VK + +C DTIEE I + K L D G NQ
Sbjct 757 AEDRVHRIGQSKQVKIFKLVCKDTIEENIFNCCQSK-LTLDNAFGGRNQ 804
> ath:AT3G57300 INO80; INO80 (INO80 ORTHOLOG); ATP binding / DNA
binding / helicase/ nucleic acid binding; K11665 DNA helicase
INO80 [EC:3.6.4.12]
Length=1507
Score = 84.0 bits (206), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 42/106 (39%), Positives = 65/106 (61%), Gaps = 4/106 (3%)
Query 12 QLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDPWWNPAAEMQ 71
+L GS T + R +++ F + +++ V L+S +AGG G+NL A + + WNP ++Q
Sbjct 1251 RLDGSSTIMDRRDMVRDFQHRSDIFVFLLSTRAGGLGINLTAADTVIFYESDWNPTLDLQ 1310
Query 72 AIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKK----QLVFDG 113
A+ RAHR+GQT+ V R IC +T+EE+IL +K QLV G
Sbjct 1311 AMDRAHRLGQTKDVTVYRLICKETVEEKILHRASQKNTVQQLVMTG 1356
> tgo:TGME49_005400 DNA repair protein RAD54, putative (EC:2.7.11.1)
Length=693
Score = 84.0 bits (206), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 51/126 (40%), Positives = 67/126 (53%), Gaps = 1/126 (0%)
Query 6 EGIHCAQLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDPWWN 65
+G +L GS R+ ++ FN L+S KAGG GLNL A+R+ L+DP WN
Sbjct 561 KGYSFLRLDGSTAVKDRTGLVKTFNESKECFAFLLSSKAGGVGLNLIGANRLVLLDPDWN 620
Query 66 PAAEMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQE-KKQLVFDGTVGASNQAMHK 124
PA + QA+ R R GQ PV R + TIEE+ILQ Q K L G VG+ Q H
Sbjct 621 PANDQQALARVWRPGQINPVYIYRLVGARTIEEKILQRQAYKATLAQVGVVGSDFQMAHI 680
Query 125 LTQEDL 130
+ +L
Sbjct 681 ILGRNL 686
> sce:YBR073W RDH54, TID1; DNA-dependent ATPase, stimulates strand
exchange by modifying the topology of double-stranded DNA;
involved in recombinational repair of DNA double-strand
breaks during mitosis and meiosis; proposed to be involved in
crossover interference (EC:5.99.-.- 3.6.1.-); K10877 DNA repair
and recombination protein RAD54B [EC:3.6.4.-]
Length=958
Score = 84.0 bits (206), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 46/122 (37%), Positives = 70/122 (57%), Gaps = 1/122 (0%)
Query 9 HCAQLTGSMTAVARSNVLYAFNNDTNLKVILISLKAGGEGLNLQVASRIYLMDPWWNPAA 68
HC +L GS+ A R +++ +FN + + L+S K+GG GLNL SR+ L D WNP+
Sbjct 708 HC-RLDGSIPAKQRDSIVTSFNRNPAIFGFLLSAKSGGVGLNLVGRSRLILFDNDWNPSV 766
Query 69 EMQAIQRAHRIGQTRPVKGVRFICTDTIEERILQLQEKKQLVFDGTVGASNQAMHKLTQE 128
++QA+ R HR GQ +P R + T I+E+ILQ Q K + +G S + + +
Sbjct 767 DLQAMSRIHRDGQKKPCFIYRLVTTGCIDEKILQRQLMKNSLSQKFLGDSEMRNKESSND 826
Query 129 DL 130
DL
Sbjct 827 DL 828
Lambda K H
0.322 0.134 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2362384368
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40