bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0759_orf2
Length=236
Score E
Sequences producing significant alignments: (Bits) Value
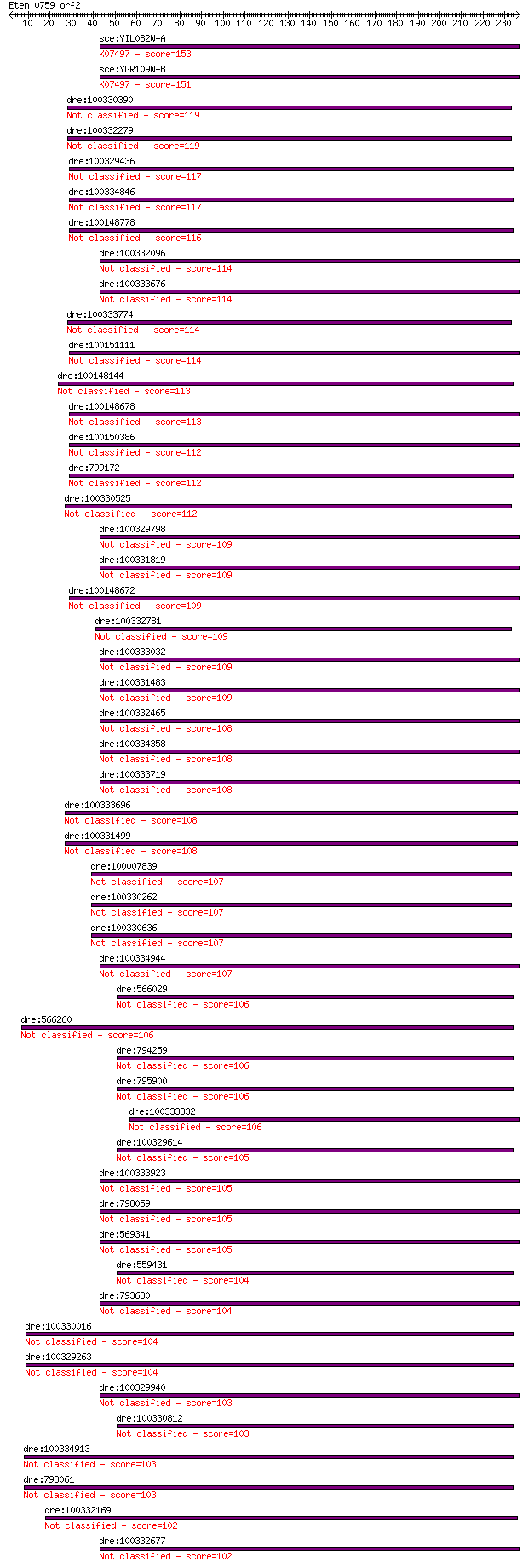
sce:YIL082W-A Retrotransposon TYA Gag and TYB Pol genes; trans... 153 6e-37
sce:YGR109W-B Retrotransposon TYA Gag and TYB Pol genes; trans... 151 2e-36
dre:100330390 RETRotransposon-like family member (retr-1)-like 119 1e-26
dre:100332279 RETRotransposon-like family member (retr-1)-like 119 1e-26
dre:100329436 RETRotransposon-like family member (retr-1)-like 117 5e-26
dre:100334846 RETRotransposon-like family member (retr-1)-like 117 5e-26
dre:100148778 RETRotransposon-like family member (retr-1)-like 116 5e-26
dre:100332096 RETRotransposon-like family member (retr-1)-like 114 2e-25
dre:100333676 RETRotransposon-like family member (retr-1)-like 114 2e-25
dre:100333774 RETRotransposon-like family member (retr-1)-like 114 3e-25
dre:100151111 RETRotransposon-like family member (retr-1)-like 114 4e-25
dre:100148144 RETRotransposon-like family member (retr-1)-like 113 6e-25
dre:100148678 RETRotransposon-like family member (retr-1)-like 113 8e-25
dre:100150386 RETRotransposon-like family member (retr-1)-like 112 8e-25
dre:799172 RETRotransposon-like family member (retr-1)-like 112 1e-24
dre:100330525 LReO_3-like 112 1e-24
dre:100329798 LReO_3-like 109 7e-24
dre:100331819 LReO_3-like 109 7e-24
dre:100148672 RETRotransposon-like family member (retr-1)-like 109 7e-24
dre:100332781 RETRotransposon-like family member (retr-1)-like 109 7e-24
dre:100333032 LReO_3-like 109 1e-23
dre:100331483 LReO_3-like 109 1e-23
dre:100332465 LReO_3-like 108 1e-23
dre:100334358 LReO_3-like 108 1e-23
dre:100333719 LReO_3-like 108 1e-23
dre:100333696 RETRotransposon-like family member (retr-1)-like 108 2e-23
dre:100331499 RETRotransposon-like family member (retr-1)-like 108 2e-23
dre:100007839 RETRotransposon-like family member (retr-1)-like 107 3e-23
dre:100330262 RETRotransposon-like family member (retr-1)-like 107 3e-23
dre:100330636 RETRotransposon-like family member (retr-1)-like 107 3e-23
dre:100334944 LReO_3-like 107 4e-23
dre:566029 LReO_3-like 106 6e-23
dre:566260 LReO_3-like 106 7e-23
dre:794259 aclyb; ATP citrate lyase b 106 8e-23
dre:795900 LReO_3-like 106 8e-23
dre:100333332 LReO_3-like 106 9e-23
dre:100329614 LReO_3-like 105 9e-23
dre:100333923 LReO_3-like 105 2e-22
dre:798059 LReO_3-like 105 2e-22
dre:569341 LReO_3-like 105 2e-22
dre:559431 si:ch211-207i1.1; si:dkey-274c14.3 104 2e-22
dre:793680 LReO_3-like 104 3e-22
dre:100330016 LReO_3-like 104 3e-22
dre:100329263 LReO_3-like 104 3e-22
dre:100329940 LReO_3-like 103 6e-22
dre:100330812 LReO_3-like 103 6e-22
dre:100334913 LReO_3-like 103 6e-22
dre:793061 LReO_3-like 103 6e-22
dre:100332169 LReO_3-like 102 8e-22
dre:100332677 LReO_3-like 102 9e-22
> sce:YIL082W-A Retrotransposon TYA Gag and TYB Pol genes; transcribed/translated
as one unit; polyprotein is processed to
make a nucleocapsid-like protein (Gag), reverse transcriptase
(RT), protease (PR), and integrase (IN); similar to retroviral
genes (EC:3.4.23.- 3.1.26.4 2.7.7.7 2.7.7.49); K07497
putative transposase
Length=1498
Score = 153 bits (386), Expect = 6e-37, Method: Compositional matrix adjust.
Identities = 76/194 (39%), Positives = 113/194 (58%), Gaps = 3/194 (1%)
Query 43 VLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCRASKSINQKP 102
V+ +H H G GH G +T A +S YYWP ++ Y+ +C C+ KS +
Sbjct 1125 VMRLYHDHTLFG---GHFGVTVTLAKISPIYYWPKLQHSIIQYIRTCVQCQLIKSHRPRL 1181
Query 103 ADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVPAKKSFTAADT 162
LLQ L I RW +S+DF+T LP T+ + ILV+VD SK AHF+ +K+ A
Sbjct 1182 HGLLQPLPIAEGRWLDISMDFVTGLPPTSNNLNMILVVVDRFSKRAHFIATRKTLDATQL 1241
Query 163 VELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQTERVN 222
++LL + YHGFP + SDR R +D + +L +R IK MSS+ HP++DGQ+ER
Sbjct 1242 IDLLFRYIFSYHGFPRTITSDRDVRMTADKYQELTKRLGIKSTMSSANHPQTDGQSERTI 1301
Query 223 RTLEQMLRTYIQSD 236
+TL ++LR Y+ ++
Sbjct 1302 QTLNRLLRAYVSTN 1315
> sce:YGR109W-B Retrotransposon TYA Gag and TYB Pol genes; transcribed/translated
as one unit; polyprotein is processed to
make a nucleocapsid-like protein (Gag), reverse transcriptase
(RT), protease (PR), and integrase (IN); similar to retroviral
genes (EC:3.4.23.- 3.1.26.4 2.7.7.7 2.7.7.49); K07497
putative transposase
Length=1547
Score = 151 bits (381), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 76/194 (39%), Positives = 112/194 (57%), Gaps = 3/194 (1%)
Query 43 VLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCRASKSINQKP 102
V+ +H H G GH G +T A +S YYWP ++ Y+ +C C+ KS +
Sbjct 1099 VMRLYHDHTLFG---GHFGVTVTLAKISPIYYWPKLQHSIIQYIRTCVQCQLIKSHRPRL 1155
Query 103 ADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVPAKKSFTAADT 162
LLQ L I RW +S+DF+T LP T+ + ILV+VD SK AHF+ +K+ A
Sbjct 1156 HGLLQPLPIAEGRWLDISMDFVTGLPPTSNNLNMILVVVDRFSKRAHFIATRKTLDATQL 1215
Query 163 VELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQTERVN 222
++LL + YHGFP + SDR R +D + +L +R IK MSS+ HP++DGQ+ER
Sbjct 1216 IDLLFRYIFSYHGFPRTITSDRDVRMTADKYQELTKRLGIKSTMSSANHPQTDGQSERTI 1275
Query 223 RTLEQMLRTYIQSD 236
+TL ++LR Y ++
Sbjct 1276 QTLNRLLRAYASTN 1289
> dre:100330390 RETRotransposon-like family member (retr-1)-like
Length=650
Score = 119 bits (298), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 70/209 (33%), Positives = 114/209 (54%), Gaps = 15/209 (7%)
Query 28 LWRICVPQFPEFLTHVLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVE 87
++++ VP F VL H HV +GH G + T+ + KH++WPG++ Y
Sbjct 10 VYQVVVPSV--FRQQVLVLAHDHVLSGHLGITK----TYHRILKHFFWPGLKQDVARYCR 63
Query 88 SCTHCRASKSINQ----KPADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDS 143
+C C+ S NQ P + + L+ PF R V VD + LP T G+ +L ++ +
Sbjct 64 TCQSCQFSGKPNQVIPPAPLNPIPILMEPFER---VIVDCVGPLPKTKAGNQFLLTIMCA 120
Query 144 LSKMAHFVPAKKSFTAADTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIK 203
++ +P +K TA ++ L + G P V+ SD+G F S+++N++ R +I+
Sbjct 121 ATRFPEAIPLRK-ITAPVIIKALT-KFFATFGLPKVVQSDQGTNFMSNVFNEVLRSLSIQ 178
Query 204 RCMSSSYHPESDGQTERVNRTLEQMLRTY 232
C+SS+YHPES G ER ++TL+ MLRTY
Sbjct 179 HCVSSAYHPESQGALERFHQTLKSMLRTY 207
> dre:100332279 RETRotransposon-like family member (retr-1)-like
Length=1088
Score = 119 bits (297), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 70/209 (33%), Positives = 114/209 (54%), Gaps = 15/209 (7%)
Query 28 LWRICVPQFPEFLTHVLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVE 87
++++ VP F VL H HV +GH G + T+ + KH++WPG++ Y
Sbjct 448 VYQVVVPSV--FRQQVLVLAHDHVLSGHLGITK----TYHRILKHFFWPGLKQDVARYCR 501
Query 88 SCTHCRASKSINQ----KPADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDS 143
+C C+ S NQ P + + L+ PF R V VD + LP T G+ +L ++ +
Sbjct 502 TCQSCQFSGKPNQVIPPAPLNPIPILMEPFER---VIVDCVGPLPKTKAGNQFLLTIMCA 558
Query 144 LSKMAHFVPAKKSFTAADTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIK 203
++ +P +K TA ++ L + G P V+ SD+G F S+++N++ R +I+
Sbjct 559 ATRFPEAIPLRK-ITAPVIIKALT-KFFATFGLPKVVQSDQGTNFMSNVFNEVLRSLSIQ 616
Query 204 RCMSSSYHPESDGQTERVNRTLEQMLRTY 232
C+SS+YHPES G ER ++TL+ MLRTY
Sbjct 617 HCVSSAYHPESQGALERFHQTLKSMLRTY 645
> dre:100329436 RETRotransposon-like family member (retr-1)-like
Length=1188
Score = 117 bits (292), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 67/205 (32%), Positives = 117/205 (57%), Gaps = 9/205 (4%)
Query 29 WRICVPQFPEFLTHVLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVES 88
++I VPQ +L ++HS+ +GH G ++ QK L + +WP M + +V++
Sbjct 808 FQIYVPQ--TLREILLEAYHSNPLSGHFGRYKTQKR----LMQVAFWPNMWRDVSDFVKN 861
Query 89 CTHCRASKSINQKPADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMA 148
CT C+ +K +KPA LQQ + W + VD + LP +T G+ +LV+VD S
Sbjct 862 CTSCQQNKPECRKPAGKLQQTEVK-EPWEMLGVDLMGPLPRSTLGNTQLLVVVDYYSHWV 920
Query 149 HFVPAKKSFTAADTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSS 208
P +K+ TA + L ++ G P L+SDRGP+F S++ LC R+ + + +++
Sbjct 921 EMFPLRKA-TAGVIAQTLRKEVLTRWGVPKFLLSDRGPQFTSEILKDLCSRWGVVQKLTT 979
Query 209 SYHPESDGQTERVNRTLEQMLRTYI 233
+YHP+++ TERVN+ ++ M+ +Y+
Sbjct 980 AYHPQTNF-TERVNQVIKVMISSYV 1003
> dre:100334846 RETRotransposon-like family member (retr-1)-like
Length=1402
Score = 117 bits (292), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 67/205 (32%), Positives = 117/205 (57%), Gaps = 9/205 (4%)
Query 29 WRICVPQFPEFLTHVLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVES 88
++I VPQ +L ++HS+ +GH G ++ QK L + +WP M + +V++
Sbjct 967 FQIYVPQ--TLREILLEAYHSNPLSGHFGRYKTQKR----LMQVAFWPNMWRDVSDFVKN 1020
Query 89 CTHCRASKSINQKPADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMA 148
CT C+ +K +KPA LQQ + W + VD + LP +T G+ +LV+VD S
Sbjct 1021 CTSCQQNKPECRKPAGKLQQTEVK-EPWEMLGVDLMGPLPRSTLGNTQLLVVVDYYSHWV 1079
Query 149 HFVPAKKSFTAADTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSS 208
P +K+ TA + L ++ G P L+SDRGP+F S++ LC R+ + + +++
Sbjct 1080 EMFPLRKA-TAGVIAQTLRKEVLTRWGVPKFLLSDRGPQFTSEILKDLCSRWGVVQKLTT 1138
Query 209 SYHPESDGQTERVNRTLEQMLRTYI 233
+YHP+++ TERVN+ ++ M+ +Y+
Sbjct 1139 AYHPQTNF-TERVNQVIKVMISSYV 1162
> dre:100148778 RETRotransposon-like family member (retr-1)-like
Length=1151
Score = 116 bits (291), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 67/205 (32%), Positives = 117/205 (57%), Gaps = 9/205 (4%)
Query 29 WRICVPQFPEFLTHVLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVES 88
++I VPQ +L ++HS+ +GH G ++ QK L + +WP M + +V++
Sbjct 763 FQIYVPQ--TLREILLEAYHSNPLSGHFGRYKTQKR----LMQVAFWPNMWRDVSDFVKN 816
Query 89 CTHCRASKSINQKPADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMA 148
CT C+ +K +KPA LQQ + W + VD + LP +T G+ +LV+VD S
Sbjct 817 CTSCQQNKPECRKPAGKLQQTEVK-EPWEMLGVDLMGPLPRSTLGNTQLLVVVDYYSHWV 875
Query 149 HFVPAKKSFTAADTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSS 208
P +K+ TA + L ++ G P L+SDRGP+F S++ LC R+ + + +++
Sbjct 876 EMFPLRKA-TAGVIAQTLRKEVLTRWGVPKFLLSDRGPQFTSEILKDLCSRWGVVQKLTT 934
Query 209 SYHPESDGQTERVNRTLEQMLRTYI 233
+YHP+++ TERVN+ ++ M+ +Y+
Sbjct 935 AYHPQTNF-TERVNQVIKVMISSYV 958
> dre:100332096 RETRotransposon-like family member (retr-1)-like
Length=456
Score = 114 bits (286), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 64/196 (32%), Positives = 107/196 (54%), Gaps = 11/196 (5%)
Query 43 VLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCRASKSINQKP 102
VL +H +GH G ++ T+ + YWPGM YV++C C+ +K N+KP
Sbjct 86 VLEWYHDTPLSGHLGIYK----TYKRIQDVAYWPGMWTDIKKYVKNCAKCQVTKWDNRKP 141
Query 103 ADLLQQLLI--PFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVPAKKSFTAA 160
A LQQ+ P W VD + +P + ++ +LV VD SK P + + TA
Sbjct 142 AGKLQQVTTSRPNEMWG---VDIMGPMPKSGKQNEYLLVFVDYFSKWVELFPMRHA-TAQ 197
Query 161 DTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQTER 220
+L ++ G P ++SDRG +F S L+ +LC ++NI ++++YHP+++ TER
Sbjct 198 TIATILRQEMLTRWGVPDFILSDRGAQFVSSLFTELCGKWNITPKLTTAYHPQTN-MTER 256
Query 221 VNRTLEQMLRTYIQSD 236
VNRTL+ M+ +++ +
Sbjct 257 VNRTLKSMIAGFVEDN 272
> dre:100333676 RETRotransposon-like family member (retr-1)-like
Length=922
Score = 114 bits (286), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 64/196 (32%), Positives = 107/196 (54%), Gaps = 11/196 (5%)
Query 43 VLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCRASKSINQKP 102
VL +H +GH G ++ T+ + YWPGM YV++C C+ +K N+KP
Sbjct 549 VLEWYHDTPLSGHLGIYK----TYKRIQDVAYWPGMWTDIKKYVKNCAKCQVTKWDNRKP 604
Query 103 ADLLQQLLI--PFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVPAKKSFTAA 160
A LQQ+ P W VD + +P + ++ +LV VD SK P + + TA
Sbjct 605 AGKLQQVTTSRPNEMWG---VDIMGPMPKSGKQNEYLLVFVDYFSKWVELFPMRHA-TAQ 660
Query 161 DTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQTER 220
+L ++ G P ++SDRG +F S L+ +LC ++NI ++++YHP+++ TER
Sbjct 661 TIATILRQEMLTRWGVPDFILSDRGAQFVSSLFTELCGKWNITPKLTTAYHPQTN-MTER 719
Query 221 VNRTLEQMLRTYIQSD 236
VNRTL+ M+ +++ +
Sbjct 720 VNRTLKSMIAGFVEDN 735
> dre:100333774 RETRotransposon-like family member (retr-1)-like
Length=1154
Score = 114 bits (285), Expect = 3e-25, Method: Composition-based stats.
Identities = 68/209 (32%), Positives = 113/209 (54%), Gaps = 15/209 (7%)
Query 28 LWRICVPQFPEFLTHVLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVE 87
++++ VP F VL H HV +GH G + T+ + KH++WPG++ Y
Sbjct 10 VYQVVVPSV--FRQQVLVLAHDHVLSGHLGITK----TYHRILKHFFWPGLKQDVARYCR 63
Query 88 SCTHCRASKSINQ----KPADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDS 143
+C C+ S NQ P + + ++ PF R V VD + LP T G+ +L ++ +
Sbjct 64 TCQSCQFSGKPNQVIPPAPLNPIPIMMEPFER---VIVDCVGPLPKTKAGNQFLLTIMCA 120
Query 144 LSKMAHFVPAKKSFTAADTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIK 203
++ +P +K TA ++ L + G P V+ SD+G F S+++N++ R +I+
Sbjct 121 ATRFPEAIPLRK-ITAPVIIKALT-KFFATFGLPKVVQSDQGTNFMSNVFNEVLRSLSIQ 178
Query 204 RCMSSSYHPESDGQTERVNRTLEQMLRTY 232
C+SS+YHPES G E ++TL+ MLRTY
Sbjct 179 HCVSSAYHPESQGALEWFHQTLKSMLRTY 207
> dre:100151111 RETRotransposon-like family member (retr-1)-like
Length=1585
Score = 114 bits (284), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 66/208 (31%), Positives = 114/208 (54%), Gaps = 9/208 (4%)
Query 29 WRICVPQFPEFLTHVLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVES 88
+R+ VP +LH +HSH +GH G ++ T+ + +WPG+ +V+
Sbjct 1198 YRVYVPN--RLRPTLLHHYHSHPLSGHHGIYK----TYKRIQAVAFWPGLWTDVKRHVKE 1251
Query 89 CTHCRASKSINQKPADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMA 148
C C+ K NQKPA LQ I R + VD I LP +T ++ +LV VD SK
Sbjct 1252 CVKCQTIKYDNQKPAGKLQS-TITSRPNQMLGVDIIGPLPRSTQQNEYLLVFVDYYSKWV 1310
Query 149 HFVPAKKSFTAADTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSS 208
F P +++ + V + L R+ G P ++SDRG +F S ++ +C ++ + + +++
Sbjct 1311 EFFPMRQANAQSVAVIFRREILTRW-GVPDFILSDRGTQFISSVFKNVCEKWGVTQKLTT 1369
Query 209 SYHPESDGQTERVNRTLEQMLRTYIQSD 236
+YHP+++ TERVNRT++ M+ +Y+ +
Sbjct 1370 AYHPQTN-MTERVNRTVKSMIASYVDDN 1396
> dre:100148144 RETRotransposon-like family member (retr-1)-like
Length=1272
Score = 113 bits (282), Expect = 6e-25, Method: Compositional matrix adjust.
Identities = 72/222 (32%), Positives = 115/222 (51%), Gaps = 20/222 (9%)
Query 24 RVHGLWRICVP----------QFPEFLT-HVLHSHHSHVTAGHRGGHRGQKMTFAALSKH 72
++ G+W VP PE LT + LH H + AGH G Q T + +
Sbjct 939 KIQGIWYRKVPLKGEGNKYQLVVPEELTQNFLHYFHDNPLAGHLG----QLKTLLKILEV 994
Query 73 YYWPGMRAYTTAYVESCTHCRASKSINQKPADLLQQLLIPFRRWAH-VSVDFITDLPLTT 131
+WP +R +YV+SC C+ K N KP+ LLQ LI H + D + P++
Sbjct 995 AWWPSVRKEVWSYVKSCKLCQQYKPSNSKPSGLLQSNLI--TEPGHTLGTDLMGPFPMSK 1052
Query 132 TGHDSILVMVDSLSKMAHFVPAKKSFTAADTVELLADRLIRYHGFPAVLISDRGPRFQSD 191
+ ILV+VD +K P + S T ++L + + G P L+SDRGP+F S
Sbjct 1053 KRNAYILVIVDYFTKWTELFPLRDSKTQK-IAKILKEEIFTRWGVPKYLVSDRGPQFTSS 1111
Query 192 LWNQLCRRFNIKRCMSSSYHPESDGQTERVNRTLEQMLRTYI 233
+ + +C+ + + ++++YHP+S+ TERVNRTL+ M+ +Y+
Sbjct 1112 ILSDVCKSWGCIQKLTTAYHPQSN-LTERVNRTLKTMIASYV 1152
> dre:100148678 RETRotransposon-like family member (retr-1)-like
Length=866
Score = 113 bits (282), Expect = 8e-25, Method: Compositional matrix adjust.
Identities = 65/208 (31%), Positives = 114/208 (54%), Gaps = 9/208 (4%)
Query 29 WRICVPQFPEFLTHVLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVES 88
+R+ VP +LH +HSH +GH G ++ T+ + +WPG+ +V+
Sbjct 583 YRVYVPN--RLRPTLLHHYHSHPLSGHHGIYK----TYKRIQAVAFWPGLWTDVKRHVKE 636
Query 89 CTHCRASKSINQKPADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMA 148
C C+ K NQKPA LQ I R + VD + LP +T ++ +LV VD SK
Sbjct 637 CVKCQTIKYDNQKPAGKLQS-TITSRPNQMLGVDIMGPLPRSTQQNEYLLVFVDYYSKWV 695
Query 149 HFVPAKKSFTAADTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSS 208
F P +++ + V + L R+ G P ++SDRG +F S ++ +C ++ + + +++
Sbjct 696 EFFPMRQANAQSVAVIFRREILTRW-GVPDFILSDRGTQFISSVFKNVCEKWGVTQKLTT 754
Query 209 SYHPESDGQTERVNRTLEQMLRTYIQSD 236
+YHP+++ TERVNRT++ M+ +Y+ +
Sbjct 755 AYHPQTN-MTERVNRTVKSMIASYVDDN 781
> dre:100150386 RETRotransposon-like family member (retr-1)-like
Length=1176
Score = 112 bits (281), Expect = 8e-25, Method: Compositional matrix adjust.
Identities = 65/208 (31%), Positives = 114/208 (54%), Gaps = 9/208 (4%)
Query 29 WRICVPQFPEFLTHVLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVES 88
+R+ VP +LH +HSH +GH G ++ T+ + +WPG+ +V+
Sbjct 762 YRVYVPN--RLRPTLLHHYHSHPLSGHHGIYK----TYKRIQAVAFWPGLWTDVKRHVKE 815
Query 89 CTHCRASKSINQKPADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMA 148
C C+ K NQKPA LQ I R + VD + LP +T ++ +LV VD SK
Sbjct 816 CVKCQTIKYDNQKPAGKLQS-TITSRPNQMLGVDIMGPLPRSTQQNEYLLVFVDYYSKWV 874
Query 149 HFVPAKKSFTAADTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSS 208
F P +++ + V + L R+ G P ++SDRG +F S ++ +C ++ + + +++
Sbjct 875 EFFPMRQANAQSVAVIFRREILTRW-GVPDFILSDRGTQFISSVFKNVCEKWGVTQKLTT 933
Query 209 SYHPESDGQTERVNRTLEQMLRTYIQSD 236
+YHP+++ TERVNRT++ M+ +Y+ +
Sbjct 934 AYHPQTN-MTERVNRTVKSMIASYVDDN 960
> dre:799172 RETRotransposon-like family member (retr-1)-like
Length=1146
Score = 112 bits (280), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 67/206 (32%), Positives = 112/206 (54%), Gaps = 11/206 (5%)
Query 29 WRICVPQFPEFLTHVLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVES 88
+++ VP+ EF + LH H + AGH G Q T + + +WP +R +YV+S
Sbjct 814 YQLVVPE--EFTQNFLHYFHDNPLAGHLG----QLKTLLKILEVAWWPSIRKEVWSYVKS 867
Query 89 CTHCRASKSINQKPADLLQQLLIPFRRWAH-VSVDFITDLPLTTTGHDSILVMVDSLSKM 147
C C+ K N KP+ LLQ LI H + D + P++ + ILV+V+ +K
Sbjct 868 CKLCQQYKPSNSKPSGLLQSNLI--TEPGHTLGTDLMGPFPMSKKRNAYILVIVEYFTKW 925
Query 148 AHFVPAKKSFTAADTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMS 207
P + S T ++L + + G P L+SDRGP+F S + + +C+ + + ++
Sbjct 926 TELFPLRDSKTQK-IAKILKEEIFTRWGVPKYLVSDRGPQFTSSILSDVCKSWGCIQKLT 984
Query 208 SSYHPESDGQTERVNRTLEQMLRTYI 233
++YHP+S+ TERVNRTL+ M+ +Y+
Sbjct 985 TAYHPQSN-LTERVNRTLKTMIASYV 1009
> dre:100330525 LReO_3-like
Length=1366
Score = 112 bits (279), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 64/210 (30%), Positives = 115/210 (54%), Gaps = 15/210 (7%)
Query 27 GLWRICVPQFPEFLTHVLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYV 86
++++ +P+ ++ +VL H H +GH G + T+ L KH++WPGM++ + Y
Sbjct 550 SVFQVVIPK--DYREYVLSVAHDHELSGHLGIRK----TYNNLLKHFFWPGMKSTVSHYC 603
Query 87 ESCTHCRASKSINQ--KPADL--LQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVD 142
+SC C+ + NQ PA L + + PF + VD + LP T +GH +L ++
Sbjct 604 QSCHACQVAGKPNQVISPAPLKPIPVMTEPFEKLV---VDCVGPLPRTKSGHSYLLTLMC 660
Query 143 SLSKMAHFVPAKKSFTAADTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNI 202
S ++ +P + S A V+ + + G P + SD+G F S ++ ++ + NI
Sbjct 661 SATRFPEAIPLR-SLKAPTIVKAIV-KFCTTFGLPKFIQSDQGSNFMSRIFRKVMKELNI 718
Query 203 KRCMSSSYHPESDGQTERVNRTLEQMLRTY 232
+ C+SS+YHP+S G ER ++TL+ ++RTY
Sbjct 719 QHCVSSAYHPQSQGVLERFHQTLKTLIRTY 748
> dre:100329798 LReO_3-like
Length=1215
Score = 109 bits (273), Expect = 7e-24, Method: Compositional matrix adjust.
Identities = 64/196 (32%), Positives = 101/196 (51%), Gaps = 11/196 (5%)
Query 43 VLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCR--ASKSINQ 100
VL H+H AGH G T + ++WPG+ Y ++C C+ + +
Sbjct 380 VLELAHTHPMAGHLGAAN----TIKRIRDRFHWPGLDGEVKRYCQACDICQRTSPQRPPP 435
Query 101 KPADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVPAKKSFTAA 160
P L + +PF R + +D I LP + GH+ ILV++D ++ +P +K+ ++A
Sbjct 436 SPLIPLPIIEVPFNR---IGMDLIGPLPKSARGHEHILVILDYATRYPEAIPLRKATSSA 492
Query 161 DTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQTER 220
EL L G PA +++D+G F S L LCR +K+ +S YHP++DG ER
Sbjct 493 IAKELFL--LCSRVGIPAEILTDQGTPFMSRLMADLCRLLKVKQIKTSVYHPQTDGLVER 550
Query 221 VNRTLEQMLRTYIQSD 236
N+TL+QMLR + D
Sbjct 551 FNKTLKQMLRRVVAED 566
> dre:100331819 LReO_3-like
Length=535
Score = 109 bits (273), Expect = 7e-24, Method: Compositional matrix adjust.
Identities = 64/196 (32%), Positives = 101/196 (51%), Gaps = 11/196 (5%)
Query 43 VLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCR--ASKSINQ 100
VL H+H AGH G T + ++WPG+ Y ++C C+ + +
Sbjct 163 VLELAHTHPMAGHLGAAN----TIKRIRDRFHWPGLDGEVKRYCQACDICQRTSPQRPPP 218
Query 101 KPADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVPAKKSFTAA 160
P L + +PF R + +D I LP + GH+ ILV++D ++ +P +K+ ++A
Sbjct 219 SPLIPLPIIEVPFNR---IGMDLIGPLPKSARGHEHILVILDYATRYPEAIPLRKATSSA 275
Query 161 DTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQTER 220
EL L G PA +++D+G F S L LCR +K+ +S YHP++DG ER
Sbjct 276 IAKELFL--LCSRVGIPAEILTDQGTPFMSRLMADLCRLLKVKQIKTSVYHPQTDGLVER 333
Query 221 VNRTLEQMLRTYIQSD 236
N+TL+QMLR + D
Sbjct 334 FNKTLKQMLRRVVAED 349
> dre:100148672 RETRotransposon-like family member (retr-1)-like
Length=1398
Score = 109 bits (273), Expect = 7e-24, Method: Compositional matrix adjust.
Identities = 64/208 (30%), Positives = 113/208 (54%), Gaps = 9/208 (4%)
Query 29 WRICVPQFPEFLTHVLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVES 88
+R+ VP +LH +HSH +GH G ++ T+ + +WPG+ +V+
Sbjct 1011 YRVYVPN--RLRPTLLHHYHSHPLSGHHGIYK----TYKRIQAVAFWPGLWTDVKRHVKE 1064
Query 89 CTHCRASKSINQKPADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMA 148
C C+ K NQKPA LQ I R + VD + LP +T ++ + V VD SK
Sbjct 1065 CVKCQTIKYDNQKPAGKLQS-TITSRPNQMLVVDIMGPLPRSTQQNEYLWVFVDYYSKWV 1123
Query 149 HFVPAKKSFTAADTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSS 208
F P +++ + V + L R+ G P ++SDRG +F S ++ +C ++ + + +++
Sbjct 1124 EFFPMRQANAQSVAVIFRREILTRW-GVPDFILSDRGTQFISSVFKNVCEKWGVTQKLTT 1182
Query 209 SYHPESDGQTERVNRTLEQMLRTYIQSD 236
+YHP+++ TERVNRT++ M+ +Y+ +
Sbjct 1183 AYHPQTN-MTERVNRTVKSMIASYVDDN 1209
> dre:100332781 RETRotransposon-like family member (retr-1)-like
Length=601
Score = 109 bits (273), Expect = 7e-24, Method: Compositional matrix adjust.
Identities = 59/196 (30%), Positives = 106/196 (54%), Gaps = 13/196 (6%)
Query 41 THVLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCRASKSINQ 100
T+VL H HV +GH G + TF +S+++YWPG+++ + + +SC C+ + NQ
Sbjct 36 TYVLKLAHEHVMSGHLGVTK----TFYRVSRYFYWPGIKSAVSEFCKSCKVCQLTGKPNQ 91
Query 101 K-PADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVPAKKSFTA 159
K P L + + + + +D + LP + +GH IL ++ + ++ VP +
Sbjct 92 KVPVAPLTPIPVINEPFERLIIDCVGPLPKSKSGHQYILTIMCAATRFPEAVPMRNIKAK 151
Query 160 ADTVELLADRLIRY---HGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDG 216
+A LI+Y G P ++ +DRG F S ++ ++ + F + +SS YHP+S G
Sbjct 152 G-----IAKELIKYCSIFGLPKIIQTDRGTNFTSKMFEKVLKEFGVNHQLSSPYHPKSQG 206
Query 217 QTERVNRTLEQMLRTY 232
ER ++TL+ MLR++
Sbjct 207 AIERFHQTLKTMLRSF 222
> dre:100333032 LReO_3-like
Length=1297
Score = 109 bits (272), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 64/196 (32%), Positives = 101/196 (51%), Gaps = 11/196 (5%)
Query 43 VLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCR--ASKSINQ 100
VL H+H AGH G T + ++WPG+ Y ++C C+ + +
Sbjct 380 VLELAHTHPMAGHLGAAN----TIKRIRDRFHWPGLDGEVKRYCQACDICQRTSPQRPPP 435
Query 101 KPADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVPAKKSFTAA 160
P L + +PF R + +D I LP + GH+ ILV++D ++ +P +K+ ++A
Sbjct 436 SPLIPLPIIEVPFNR---IGMDLIGPLPKSARGHEHILVILDYATRYPEAIPLRKATSSA 492
Query 161 DTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQTER 220
EL L G PA +++D+G F S L LCR +K+ +S YHP++DG ER
Sbjct 493 IAKELFL--LCSRVGIPAEILTDQGTPFMSRLMADLCRLLKVKQIKTSVYHPQTDGLVER 550
Query 221 VNRTLEQMLRTYIQSD 236
N+TL+QMLR + D
Sbjct 551 FNKTLKQMLRRVVAED 566
> dre:100331483 LReO_3-like
Length=1352
Score = 109 bits (272), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 64/196 (32%), Positives = 101/196 (51%), Gaps = 11/196 (5%)
Query 43 VLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCR--ASKSINQ 100
VL H+H AGH G T + ++WPG+ Y ++C C+ + +
Sbjct 400 VLELAHTHPMAGHLGAAN----TIKRIRDRFHWPGLDGEVKRYCQACDICQRTSPQRPPP 455
Query 101 KPADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVPAKKSFTAA 160
P L + +PF R + +D I LP + GH+ ILV++D ++ +P +K+ ++A
Sbjct 456 SPLIPLPIIEVPFNR---IGMDLIGPLPKSARGHEHILVILDYATRYPEAIPLRKATSSA 512
Query 161 DTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQTER 220
EL L G PA +++D+G F S L LCR +K+ +S YHP++DG ER
Sbjct 513 IAKELFL--LCSRVGIPAEILTDQGTPFMSRLMADLCRLLKVKQIKTSVYHPQTDGLVER 570
Query 221 VNRTLEQMLRTYIQSD 236
N+TL+QMLR + D
Sbjct 571 FNKTLKQMLRRVVAED 586
> dre:100332465 LReO_3-like
Length=1304
Score = 108 bits (271), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 64/196 (32%), Positives = 101/196 (51%), Gaps = 11/196 (5%)
Query 43 VLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCR--ASKSINQ 100
VL H+H AGH G T + ++WPG+ Y ++C C+ + +
Sbjct 494 VLELAHTHPMAGHLGAAN----TIKRIRDRFHWPGLDGEVKRYCQACDICQRTSPQRPPP 549
Query 101 KPADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVPAKKSFTAA 160
P L + +PF R + +D I LP + GH+ ILV++D ++ +P +K+ ++A
Sbjct 550 SPLIPLPIIEVPFNR---IGMDLIGPLPKSARGHEHILVILDYATRYPEAIPLRKATSSA 606
Query 161 DTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQTER 220
EL L G PA +++D+G F S L LCR +K+ +S YHP++DG ER
Sbjct 607 IAKELFL--LCSRVGIPAEILTDQGTPFMSRLMADLCRLLEVKQIKTSVYHPQTDGLVER 664
Query 221 VNRTLEQMLRTYIQSD 236
N+TL+QMLR + D
Sbjct 665 FNKTLKQMLRRVVAED 680
> dre:100334358 LReO_3-like
Length=1188
Score = 108 bits (271), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 64/196 (32%), Positives = 101/196 (51%), Gaps = 11/196 (5%)
Query 43 VLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCR--ASKSINQ 100
VL H+H AGH G T + ++WPG+ Y ++C C+ + +
Sbjct 370 VLELAHTHPMAGHLGAAN----TIKRIRDRFHWPGLDGEVKRYCQACDICQRTSPQRPPP 425
Query 101 KPADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVPAKKSFTAA 160
P L + +PF R + +D I LP + GH+ ILV++D ++ +P +K+ ++A
Sbjct 426 SPLIPLPIIEVPFNR---IGMDLIGPLPKSARGHEHILVILDYATRYPKAIPLRKATSSA 482
Query 161 DTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQTER 220
EL L G PA +++D+G F S L LCR +K+ +S YHP++DG ER
Sbjct 483 IAKELFL--LCSRVGIPAEILTDQGTPFMSRLMADLCRLLKVKQIKTSVYHPQTDGLVER 540
Query 221 VNRTLEQMLRTYIQSD 236
N+TL+QMLR + D
Sbjct 541 FNKTLKQMLRRVVAED 556
> dre:100333719 LReO_3-like
Length=1121
Score = 108 bits (271), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 64/196 (32%), Positives = 101/196 (51%), Gaps = 11/196 (5%)
Query 43 VLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCR--ASKSINQ 100
VL H+H AGH G T + ++WPG+ Y ++C C+ + +
Sbjct 372 VLELAHTHPMAGHLGAAN----TIKRIRDRFHWPGLDGEVKRYCQACDICQRTSPQRPPP 427
Query 101 KPADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVPAKKSFTAA 160
P L + +PF R + +D I LP + GH+ ILV++D ++ +P +K+ ++A
Sbjct 428 SPLIPLPIIEVPFNR---IGMDLIGPLPKSARGHEHILVILDYATRYPEAIPLRKATSSA 484
Query 161 DTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQTER 220
EL L G PA +++D+G F S L LCR +K+ +S YHP++DG ER
Sbjct 485 IAKELFL--LCSRVGIPAEILTDQGTPFMSRLMADLCRLLKVKQIKTSVYHPQTDGLVER 542
Query 221 VNRTLEQMLRTYIQSD 236
N+TL+QMLR + D
Sbjct 543 FNKTLKQMLRRVVAED 558
> dre:100333696 RETRotransposon-like family member (retr-1)-like
Length=1036
Score = 108 bits (269), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 58/210 (27%), Positives = 110/210 (52%), Gaps = 9/210 (4%)
Query 27 GLWRICVPQFPEFLTHVLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYV 86
++++ +P + + VL H H +GH G ++ TF +S++++WPG+++ + +
Sbjct 285 AIYQVVLPS--TYRSFVLQLAHEHPLSGHLGVNK----TFKRISRYFFWPGLKSSVSKFC 338
Query 87 ESCTHCRASKSINQK-PADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLS 145
+ C C+ + NQ P+ L + + + +D + LP + +GH IL ++ + +
Sbjct 339 KQCHVCQIAGKPNQTIPSAPLHPIPAVGEPFERLILDCVGPLPKSKSGHQYILTLMCTAT 398
Query 146 KMAHFVPAKKSFTAADTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRC 205
+ +P + A E++ + G P V+ +DRG F S L+ Q+C+ +
Sbjct 399 RFPEAIPLRSLKAKAIVKEVV--KFCSTFGLPKVIQTDRGTNFTSKLFTQICKELGVTNQ 456
Query 206 MSSSYHPESDGQTERVNRTLEQMLRTYIQS 235
+SS+YHPES G ER ++TL+ MLR Y S
Sbjct 457 LSSAYHPESQGALERFHQTLKSMLRAYCTS 486
> dre:100331499 RETRotransposon-like family member (retr-1)-like
Length=1264
Score = 108 bits (269), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 60/213 (28%), Positives = 111/213 (52%), Gaps = 15/213 (7%)
Query 27 GLWRICVPQFPEFLTHVLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYV 86
++++ +P + + VL H H +GH G ++ TF +S++++WPG+++ + +
Sbjct 430 AIYQVVLPS--TYQSFVLQLAHEHPLSGHLGVNK----TFKRISRYFFWPGLKSSVSKFC 483
Query 87 ESCTHCRASKSINQ----KPADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVD 142
+ C C+ + NQ P + + PF R + +D + LP + +GH IL ++
Sbjct 484 KQCHVCQIAGKPNQTIPSAPLHPIPAVGEPFER---LILDCVGPLPKSKSGHQYILTLMC 540
Query 143 SLSKMAHFVPAKKSFTAADTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNI 202
+ ++ +P + A E++ + G P V+ +DRG F S L+ Q+C+ +
Sbjct 541 TATRFPEAIPLRSLKAKAIVKEVV--KFCSTFGLPKVIQTDRGTNFTSKLFTQICKELGV 598
Query 203 KRCMSSSYHPESDGQTERVNRTLEQMLRTYIQS 235
+SS+YHPES G ER ++TL+ MLR Y S
Sbjct 599 TNQLSSAYHPESQGALERFHQTLKSMLRAYCTS 631
> dre:100007839 RETRotransposon-like family member (retr-1)-like
Length=731
Score = 107 bits (268), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 60/195 (30%), Positives = 104/195 (53%), Gaps = 7/195 (3%)
Query 39 FLTHVLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCRASKSI 98
+ + +L H HV +GH G + T+ + ++++WPG+++ +AY SC C+ +
Sbjct 156 YRSQILALAHDHVFSGHLGVRK----TYDRILRYFFWPGLKSDVSAYCRSCHSCQLAGKP 211
Query 99 NQK-PADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVPAKKSF 157
NQ P L + I + + +D + LP + GH IL ++ + ++ VP +
Sbjct 212 NQVIPPAPLHPIPITSEPFERILIDCVGPLPKSKAGHQYILTIMCAATRYPEAVPMRNIK 271
Query 158 TAADTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQ 217
A EL+ + G P V+ +D+G F S L++Q+ R +K +SS+YHPES G
Sbjct 272 AKAVVRELI--KFCSTFGLPKVIQTDQGSNFTSTLFSQVLRELTVKHQLSSAYHPESQGV 329
Query 218 TERVNRTLEQMLRTY 232
ER ++TL+ MLRT+
Sbjct 330 LERFHQTLKSMLRTF 344
> dre:100330262 RETRotransposon-like family member (retr-1)-like
Length=728
Score = 107 bits (268), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 60/195 (30%), Positives = 104/195 (53%), Gaps = 7/195 (3%)
Query 39 FLTHVLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCRASKSI 98
+ + +L H HV +GH G + T+ + ++++WPG+++ +AY SC C+ +
Sbjct 156 YRSQILALAHDHVFSGHLGVRK----TYDRILRYFFWPGLKSDVSAYCRSCHSCQLAGKP 211
Query 99 NQK-PADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVPAKKSF 157
NQ P L + I + + +D + LP + GH IL ++ + ++ VP +
Sbjct 212 NQVIPPAPLHPIPITSEPFERILIDCVGPLPKSKAGHQYILTIMCAATRYPEAVPMRNIK 271
Query 158 TAADTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQ 217
A EL+ + G P V+ +D+G F S L++Q+ R +K +SS+YHPES G
Sbjct 272 AKAVVRELI--KFCSTFGLPKVIQTDQGSNFTSTLFSQVLRELTVKHQLSSAYHPESQGV 329
Query 218 TERVNRTLEQMLRTY 232
ER ++TL+ MLRT+
Sbjct 330 LERFHQTLKSMLRTF 344
> dre:100330636 RETRotransposon-like family member (retr-1)-like
Length=1406
Score = 107 bits (268), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 60/195 (30%), Positives = 104/195 (53%), Gaps = 7/195 (3%)
Query 39 FLTHVLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCRASKSI 98
+ + +L H HV +GH G + T+ + ++++WPG+++ +AY SC C+ +
Sbjct 542 YRSQILALAHDHVFSGHLGVRK----TYDRILRYFFWPGLKSDVSAYCRSCHSCQLAGKP 597
Query 99 NQK-PADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVPAKKSF 157
NQ P L + I + + +D + LP + GH IL ++ + ++ VP +
Sbjct 598 NQVIPPAPLHPIPITSEPFERILIDCVGPLPKSKAGHQYILTIMCAATRYPEAVPMRNIK 657
Query 158 TAADTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQ 217
A EL+ + G P V+ +D+G F S L++Q+ R +K +SS+YHPES G
Sbjct 658 AKAVVRELI--KFCSTFGLPKVIQTDQGSNFTSTLFSQVLRELTVKHQLSSAYHPESQGV 715
Query 218 TERVNRTLEQMLRTY 232
ER ++TL+ MLRT+
Sbjct 716 LERFHQTLKSMLRTF 730
> dre:100334944 LReO_3-like
Length=1181
Score = 107 bits (267), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 64/196 (32%), Positives = 101/196 (51%), Gaps = 11/196 (5%)
Query 43 VLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCR--ASKSINQ 100
VL H+H AGH G K + ++WPG+ Y ++C C+ + +
Sbjct 380 VLELAHTHPMAGHLGAANRIKR----IRDRFHWPGLDGEVKRYCQACDICQRTSPQRPPP 435
Query 101 KPADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVPAKKSFTAA 160
P L + +PF R + +D I LP + GH+ ILV++D ++ +P +K+ ++A
Sbjct 436 SPLIPLPIIEVPFNR---IGMDLIGPLPKSARGHEHILVILDYATRYPEAIPLRKATSSA 492
Query 161 DTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQTER 220
EL L G PA +++D+G F S L LCR +K+ +S YHP++DG ER
Sbjct 493 IAKELFL--LCSRVGIPAEILTDQGTPFMSRLMADLCRLLKVKQIKTSVYHPQTDGLVER 550
Query 221 VNRTLEQMLRTYIQSD 236
N+TL+QMLR + D
Sbjct 551 FNKTLKQMLRRVVAED 566
> dre:566029 LReO_3-like
Length=1490
Score = 106 bits (265), Expect = 6e-23, Method: Compositional matrix adjust.
Identities = 60/191 (31%), Positives = 102/191 (53%), Gaps = 16/191 (8%)
Query 51 VTAGHR---GGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCR--ASKSINQKPADL 105
+T GH GH G++ T A + KH+YWPG+ + Y ++C C+ +S + P
Sbjct 635 LTLGHSIPWAGHLGKRKTIARIRKHFYWPGLNSEVAQYCKTCPECQKVSSDCPRRVPLQP 694
Query 106 LQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVP---AKKSFTAADT 162
L + PF R + +D + + + +G+ +LV+ D ++ P K + A
Sbjct 695 LPLIGTPFER---LGMDIVGPVEKSRSGNRFMLVVTDYATRYPEVFPLRSVKAKYVATCL 751
Query 163 VELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQTERVN 222
V+L + GFP+ +++D+G F S+L Q+ R IK ++ YHP++DG TER N
Sbjct 752 VQLFS-----RVGFPSEILTDQGTNFMSNLLKQVYRLLGIKSLRTTPYHPQTDGLTERFN 806
Query 223 RTLEQMLRTYI 233
+TL+QMLR ++
Sbjct 807 QTLKQMLRKFV 817
> dre:566260 LReO_3-like
Length=1496
Score = 106 bits (265), Expect = 7e-23, Method: Compositional matrix adjust.
Identities = 69/230 (30%), Positives = 116/230 (50%), Gaps = 16/230 (6%)
Query 7 FCNRQFTFRYVEPYLHIRVHGLWRICVPQFPEFLTHVLHSHHSHVTAGHRGGHRGQKMTF 66
CN + L++R + + R+ VP L VLH H+ AGH G Q+ T+
Sbjct 601 LCNEVYVVE--NGVLYVRTNDVLRLVVPSCCRPL--VLHLAHTVPWAGHLG----QQKTY 652
Query 67 AALSKHYYWPGMRAYTTAYVESCTHCRASKSINQKPADLLQQLLI---PFRRWAHVSVDF 123
A +S +YWP + + ++C C+ + +++Q+ LQ L + PFRR +++D
Sbjct 653 ARISSRFYWPTLYTDVQTHCKTCAVCQKTSAVSQRGRAPLQPLPVISAPFRR---IAMDI 709
Query 124 ITDLPLTTTGHDSILVMVDSLSKMAHFVPAKKSFTAADTVELLADRLIRYHGFPAVLISD 183
+ L ++ GH ILV+ D ++ P +S T + L R G P +++D
Sbjct 710 VGPLEKSSAGHRYILVVSDYATRYPEAYPL-RSITTPKIIHALIQLFSRV-GIPEEILTD 767
Query 184 RGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQTERVNRTLEQMLRTYI 233
+G F S L QL ++ I ++ YHP++DG ER N+TL+ MLR ++
Sbjct 768 QGTNFTSRLMGQLHKQMGITAIRTTPYHPQTDGLVERFNQTLKNMLRKFV 817
> dre:794259 aclyb; ATP citrate lyase b
Length=2571
Score = 106 bits (264), Expect = 8e-23, Method: Compositional matrix adjust.
Identities = 61/191 (31%), Positives = 104/191 (54%), Gaps = 16/191 (8%)
Query 51 VTAGHR---GGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCR--ASKSINQKPADL 105
+T GH GH G++ T A + KH+YWPG+ + Y ++C C+ +S + P
Sbjct 1107 LTLGHSIPWAGHLGKRKTIARIRKHFYWPGLNSEVAQYCKTCPECQKVSSDCPRRVPLQP 1166
Query 106 LQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVP---AKKSFTAADT 162
L + PF R + +D + + + +G+ +LV+ D ++ P K + A
Sbjct 1167 LPLIGTPFER---LGMDIVGPVEKSRSGNRFMLVVTDYATRYPEVFPLRSVKAKYVATCL 1223
Query 163 VELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQTERVN 222
V+L + R+ GFP+ +++D+G F S+L Q+ R IK ++ YHP++DG TER N
Sbjct 1224 VQLFS-RV----GFPSEILTDQGTNFMSNLLKQVYRLLGIKSLRTTPYHPQTDGLTERFN 1278
Query 223 RTLEQMLRTYI 233
+TL+QMLR ++
Sbjct 1279 QTLKQMLRKFV 1289
> dre:795900 LReO_3-like
Length=1490
Score = 106 bits (264), Expect = 8e-23, Method: Compositional matrix adjust.
Identities = 60/188 (31%), Positives = 100/188 (53%), Gaps = 10/188 (5%)
Query 51 VTAGHR---GGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCR--ASKSINQKPADL 105
+T GH GH G++ T A + KH+YWPG+ + Y ++C C+ +S + P
Sbjct 635 LTLGHSIPWAGHLGKRKTIARIRKHFYWPGLNSEVAQYCKTCPECQKVSSDCPRRVPLQP 694
Query 106 LQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVPAKKSFTAADTVEL 165
L + PF R + +D + + + +G+ +LV+ D ++ P + S A
Sbjct 695 LPLIGTPFER---LGMDIVGPVEKSRSGNRFMLVVTDYATRYPEVFPLR-SVKAKHVATC 750
Query 166 LADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQTERVNRTL 225
L R GFP+ +++D+G F S+L Q+ R IK ++ YHP++DG TER N+TL
Sbjct 751 LVQLFSRV-GFPSEILTDQGTNFMSNLLKQVYRLLGIKSLRTTPYHPQTDGLTERFNQTL 809
Query 226 EQMLRTYI 233
+QMLR ++
Sbjct 810 KQMLRKFV 817
> dre:100333332 LReO_3-like
Length=963
Score = 106 bits (264), Expect = 9e-23, Method: Compositional matrix adjust.
Identities = 57/182 (31%), Positives = 94/182 (51%), Gaps = 7/182 (3%)
Query 57 GGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCRASKSINQKPADLLQQLLI--PFR 114
GH G T + ++WPG+ Y ++C C+ P+ L+ +I PF
Sbjct 296 AGHLGAANTVKRIRDRFHWPGLDGEVKRYCQACDICQRMSPQRPPPSPLITLPIIDVPFT 355
Query 115 RWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVPAKKSFTAADTVELLADRLIRYH 174
R + +D + LP + GH+ ILV++D ++ +P +K+ ++A EL L
Sbjct 356 R---IGMDLVGPLPKSARGHEHILVILDYATRYPEAIPLRKATSSAIAKELFL--LCSRV 410
Query 175 GFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQTERVNRTLEQMLRTYIQ 234
G PA +++D+G F S L LC +K+ +S YHP++DG ER N+TL+QML+ +
Sbjct 411 GIPAEILTDQGTPFMSRLMADLCHLLKVKQIKTSVYHPQTDGLVERFNKTLKQMLQRVVA 470
Query 235 SD 236
D
Sbjct 471 ED 472
> dre:100329614 LReO_3-like
Length=1490
Score = 105 bits (263), Expect = 9e-23, Method: Compositional matrix adjust.
Identities = 60/191 (31%), Positives = 101/191 (52%), Gaps = 16/191 (8%)
Query 51 VTAGHR---GGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCR--ASKSINQKPADL 105
+T GH GH G++ T A + KH+YWPG+ + Y ++C C+ +S + P
Sbjct 635 LTLGHSIPWAGHLGKRKTIARIRKHFYWPGLNSEVAQYCKTCPECQKVSSDCPRRVPLQP 694
Query 106 LQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVP---AKKSFTAADT 162
L + PF R + +D + + + G+ +LV+ D ++ P K + A
Sbjct 695 LPLIGTPFER---LGMDLVGPVEKSRLGNRFMLVVTDYATRYPEVFPLRSVKAKYVATCL 751
Query 163 VELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQTERVN 222
V+L + GFP+ +++D+G F S+L Q+ R IK ++ YHP++DG TER N
Sbjct 752 VQLFS-----RVGFPSEILTDQGTNFMSNLLKQVYRLLGIKSLRTTPYHPQTDGLTERFN 806
Query 223 RTLEQMLRTYI 233
+TL+QMLR ++
Sbjct 807 QTLKQMLRKFV 817
> dre:100333923 LReO_3-like
Length=1299
Score = 105 bits (261), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 65/198 (32%), Positives = 103/198 (52%), Gaps = 15/198 (7%)
Query 43 VLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCRASKSINQKP 102
+L HSH AGH G T + ++WPG+ A + ++C C+ + P
Sbjct 334 ILELAHSHPMAGHLG----VANTVQRIRDRFHWPGLEADVKRFCQACPTCQRTSPRTPPP 389
Query 103 ADLLQQLL--IPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVPAKKSFTAA 160
+ L+ + +PF R + +D + LP + GH+ ILV+VD ++ VP +K+ A
Sbjct 390 SPLIPLPIIEVPFER---IGMDLVGPLPKSARGHEHILVIVDYATRYPEAVPLRKATAKA 446
Query 161 DTVEL--LADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQT 218
EL L+ R+ G PA +++D+G F S L LCR +K+ ++ YHP++DG
Sbjct 447 IAQELFLLSSRV----GIPAEILTDQGTPFMSRLMADLCRLLKVKQLRTTVYHPQTDGLV 502
Query 219 ERVNRTLEQMLRTYIQSD 236
ER N+TL+QMLR D
Sbjct 503 ERFNQTLKQMLRRVAAED 520
> dre:798059 LReO_3-like
Length=1210
Score = 105 bits (261), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 65/198 (32%), Positives = 103/198 (52%), Gaps = 15/198 (7%)
Query 43 VLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCRASKSINQKP 102
+L HSH AGH G T + ++WPG+ A + ++C C+ + P
Sbjct 378 ILELAHSHPMAGHLG----VANTVQRIRDRFHWPGLEADVKRFCQACPTCQQTSPRTPPP 433
Query 103 ADLLQQLL--IPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVPAKKSFTAA 160
+ L+ + +PF R + +D + LP + GH+ ILV+VD ++ VP +K+ A
Sbjct 434 SPLIPLPIIEVPFER---IGMDLVGPLPKSARGHEHILVIVDYATRYPEAVPLRKATAKA 490
Query 161 DTVEL--LADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQT 218
EL L+ R+ G PA +++D+G F S L LCR +K+ ++ YHP++DG
Sbjct 491 IAQELFLLSSRV----GIPAEILTDQGTPFMSRLMADLCRLLKVKQLRTTVYHPQTDGLV 546
Query 219 ERVNRTLEQMLRTYIQSD 236
ER N+TL+QMLR D
Sbjct 547 ERFNQTLKQMLRRVAAED 564
> dre:569341 LReO_3-like
Length=1379
Score = 105 bits (261), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 65/198 (32%), Positives = 103/198 (52%), Gaps = 15/198 (7%)
Query 43 VLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCRASKSINQKP 102
+L HSH H GH G T + ++WPG+ A + ++C C+ + P
Sbjct 512 ILELAHSH----HMAGHLGVANTVQRIRDCFHWPGLEADVKRFCQACPTCQRTSPRTPPP 567
Query 103 ADLLQQLL--IPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVPAKKSFTAA 160
+ L+ + +PF R + +D + LP + GH+ ILV+VD ++ VP +K+ A
Sbjct 568 SPLIPLPIIKVPFER---IGMDLVGPLPKSARGHEHILVIVDYATRYPEAVPLRKATAKA 624
Query 161 DTVEL--LADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQT 218
EL L+ R+ G PA +++D+G F S L LCR +K+ ++ YHP++DG
Sbjct 625 IAQELFLLSSRV----GIPAEILTDQGTPFMSRLMADLCRLLKVKQLRTTVYHPQTDGLV 680
Query 219 ERVNRTLEQMLRTYIQSD 236
ER N+TL+QMLR D
Sbjct 681 ERFNQTLKQMLRRVAVED 698
> dre:559431 si:ch211-207i1.1; si:dkey-274c14.3
Length=9154
Score = 104 bits (260), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 60/191 (31%), Positives = 102/191 (53%), Gaps = 16/191 (8%)
Query 51 VTAGHR---GGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCR--ASKSINQKPADL 105
+T GH GH G++ T A + KH+YWPG+ + Y ++C C+ +S + P
Sbjct 635 LTLGHSIPWAGHLGKRKTIARIRKHFYWPGLNSVVAQYCKTCPECQKVSSDCPRRVPLQP 694
Query 106 LQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVP---AKKSFTAADT 162
L + PF R + +D + + + +G+ +LV+ D ++ P K + A
Sbjct 695 LPLIGTPFER---LGMDIVGPVEKSRSGNRFMLVVTDYATRYPEVFPLRSVKAKYVATCL 751
Query 163 VELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQTERVN 222
V+L + GFP+ +++D+G F S+L Q+ R IK ++ YHP++DG TER N
Sbjct 752 VQLFS-----RVGFPSEILTDQGTNFMSNLLKQVYRLLGIKSLRTTPYHPQTDGLTERFN 806
Query 223 RTLEQMLRTYI 233
+TL+QMLR ++
Sbjct 807 QTLKQMLRKFV 817
> dre:793680 LReO_3-like
Length=1368
Score = 104 bits (260), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 65/198 (32%), Positives = 103/198 (52%), Gaps = 15/198 (7%)
Query 43 VLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCRASKSINQKP 102
+L HSH AGH G T + ++WPG+ A + ++C C+ + P
Sbjct 403 ILELAHSHPMAGHLG----VANTVQRIRDRFHWPGLEADVKWFCQACPTCQRTSPRTPPP 458
Query 103 ADLLQQLL--IPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVPAKKSFTAA 160
+ L+ + +PF R + +D + LP + GH+ ILV+VD ++ VP +K+ A
Sbjct 459 SPLIPLPIIEVPFER---IGMDLVGPLPKSARGHEHILVIVDYATRYPEAVPLRKATAKA 515
Query 161 DTVEL--LADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQT 218
EL L+ R+ G PA +++D+G F S L LCR +K+ ++ YHP++DG
Sbjct 516 IAQELFLLSSRV----GIPAEILTDQGTPFMSRLMADLCRLLKVKQLRTTVYHPQTDGLV 571
Query 219 ERVNRTLEQMLRTYIQSD 236
ER N+TL+QMLR D
Sbjct 572 ERFNQTLKQMLRRVAAED 589
> dre:100330016 LReO_3-like
Length=1476
Score = 104 bits (259), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 66/227 (29%), Positives = 116/227 (51%), Gaps = 13/227 (5%)
Query 9 NRQFTFRYVEPYLHIRVHGLWRICVPQFPEFLTHVLHSHHSHVTAGHRGGHRGQKMTFAA 68
++ TF ++ L+ ++ ++ VPQ + VLH HS AGH G ++ T A
Sbjct 594 DKTVTFVRIKGILYRQIGPRRQLVVPQCVREV--VLHLSHSVPWAGHLGKNK----TIAR 647
Query 69 LSKHYYWPGMRAYTTAYVESCTHCR--ASKSINQKPADLLQQLLIPFRRWAHVSVDFITD 126
+ +++YWPG+ + +SC C+ + + + P L + PF R + +D +
Sbjct 648 IKRYFYWPGLEGDVAQFCKSCPVCQKVSLQRPRKAPLQPLPIITTPFER---LGMDVVGP 704
Query 127 LPLTTTGHDSILVMVDSLSKMAHFVPAKKSFTAADTVELLADRLIRYHGFPAVLISDRGP 186
L + +G+ +LV+ D ++ P + A +V +L GFP +++D+G
Sbjct 705 LEKSRSGNRFMLVITDYATRYPEVFPLRS--VKAKSVATSLVQLFSRVGFPMEILTDQGT 762
Query 187 RFQSDLWNQLCRRFNIKRCMSSSYHPESDGQTERVNRTLEQMLRTYI 233
F S L Q+ R IK ++ YHP++DG TER N+TL+QMLR ++
Sbjct 763 NFMSTLLKQVYRLLGIKSIRTTPYHPQTDGLTERFNQTLKQMLRKFV 809
> dre:100329263 LReO_3-like
Length=1476
Score = 104 bits (259), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 66/227 (29%), Positives = 116/227 (51%), Gaps = 13/227 (5%)
Query 9 NRQFTFRYVEPYLHIRVHGLWRICVPQFPEFLTHVLHSHHSHVTAGHRGGHRGQKMTFAA 68
++ TF ++ L+ ++ ++ VPQ + VLH HS AGH G ++ T A
Sbjct 594 DKTVTFVRIKGILYRQIGPRRQLVVPQCVREV--VLHLSHSVPWAGHLGKNK----TIAR 647
Query 69 LSKHYYWPGMRAYTTAYVESCTHCR--ASKSINQKPADLLQQLLIPFRRWAHVSVDFITD 126
+ +++YWPG+ + +SC C+ + + + P L + PF R + +D +
Sbjct 648 IKRYFYWPGLEGDVAQFCKSCPVCQKVSLQRPRKAPLQPLPIITTPFER---LGMDVVGP 704
Query 127 LPLTTTGHDSILVMVDSLSKMAHFVPAKKSFTAADTVELLADRLIRYHGFPAVLISDRGP 186
L + +G+ +LV+ D ++ P + A +V +L GFP +++D+G
Sbjct 705 LEKSRSGNRFMLVITDYATRYPEVFPLRS--VKAKSVATSLVQLFSRVGFPMEILTDQGT 762
Query 187 RFQSDLWNQLCRRFNIKRCMSSSYHPESDGQTERVNRTLEQMLRTYI 233
F S L Q+ R IK ++ YHP++DG TER N+TL+QMLR ++
Sbjct 763 NFMSTLLKQVYRLLGIKSIRTTPYHPQTDGLTERFNQTLKQMLRKFV 809
> dre:100329940 LReO_3-like
Length=1643
Score = 103 bits (257), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 62/196 (31%), Positives = 100/196 (51%), Gaps = 11/196 (5%)
Query 43 VLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCR--ASKSINQ 100
VL H+H AGH G T + ++WPG+ Y ++C C+ + +
Sbjct 494 VLELAHTHPMAGHLGAAN----TIKRIRDRFHWPGLDGEVKRYCQACDICQRTSPQRPPP 549
Query 101 KPADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVPAKKSFTAA 160
P L + +PF R + +D I LP + GH+ ILV++D ++ +P +K+ ++A
Sbjct 550 SPLIPLPIIEVPFNR---IGMDLIGPLPKSARGHEHILVILDYATRYPEAIPLRKATSSA 606
Query 161 DTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQTER 220
EL L G A +++D+G F S L LCR +K+ +S YHP++DG +R
Sbjct 607 IAKELFL--LCSRVGILAEILTDQGTPFMSRLMADLCRLLKVKQIKTSVYHPQTDGLVKR 664
Query 221 VNRTLEQMLRTYIQSD 236
N+TL+QMLR + D
Sbjct 665 FNKTLKQMLRRVVAED 680
> dre:100330812 LReO_3-like
Length=1335
Score = 103 bits (256), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 59/191 (30%), Positives = 101/191 (52%), Gaps = 16/191 (8%)
Query 51 VTAGHR---GGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCR--ASKSINQKPADL 105
+T GH GH G++ T A + KH+YWPG+ + Y ++ C+ +S + P
Sbjct 635 LTLGHSIPWAGHLGKRKTIARIRKHFYWPGLNSEVAQYCKTSPECQKVSSDCPRRVPLQP 694
Query 106 LQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVP---AKKSFTAADT 162
L + PF R + +D + + + +G+ +LV+ D ++ P K + A
Sbjct 695 LPLIGTPFER---LGMDIVGPVEKSRSGNRFMLVVTDYATRYPEVFPLRSVKAKYVATCL 751
Query 163 VELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQTERVN 222
V+L + GFP+ +++D+G F S+L Q+ R IK ++ YHP++DG TER N
Sbjct 752 VQLFS-----RVGFPSEILTDQGTNFMSNLLKQVYRMLGIKSLRTTPYHPQTDGLTERFN 806
Query 223 RTLEQMLRTYI 233
+TL+QMLR ++
Sbjct 807 QTLKQMLRKFV 817
> dre:100334913 LReO_3-like
Length=886
Score = 103 bits (256), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 72/229 (31%), Positives = 116/229 (50%), Gaps = 18/229 (7%)
Query 8 CNRQFTFRYVEPYLHIRVHGLWRICVPQFPEFLTHVLHSHHSHVTAGHRGGHRGQKMTFA 67
C R F E L+ ++ + ++ VP+ E VLH HS AGH G + T
Sbjct 528 CER---FMISENILYRQIGSVKQLIVPK--EARELVLHLSHSIPWAGHLGVRK----TIE 578
Query 68 ALSKHYYWPGMRAYTTAYVESCTHCRASKSINQKPADLLQQLLI---PFRRWAHVSVDFI 124
+ K++YWPG++A Y +SC C+ S++ P LQ L + PF R + +D +
Sbjct 579 RIKKYFYWPGLKADVVQYCKSCPECQLV-SLHHPPRVPLQPLPLISTPFER---LGMDIV 634
Query 125 TDLPLTTTGHDSILVMVDSLSKMAHFVPAKKSFTAADTVELLADRLIRYHGFPAVLISDR 184
+ ++ G+ +LV+ D ++ P K S A + L R G P +++D+
Sbjct 635 GPVEKSSLGNRFLLVITDYATRYPEVFPLK-SMKAKNVATCLIQFCSRV-GLPREILTDQ 692
Query 185 GPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQTERVNRTLEQMLRTYI 233
G F S L Q+ + IK ++ YHP++DG TER N+TL+QMLR ++
Sbjct 693 GTNFMSTLLKQVYQLLGIKSLRTTPYHPQTDGLTERFNQTLKQMLRKFV 741
> dre:793061 LReO_3-like
Length=1490
Score = 103 bits (256), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 72/229 (31%), Positives = 116/229 (50%), Gaps = 18/229 (7%)
Query 8 CNRQFTFRYVEPYLHIRVHGLWRICVPQFPEFLTHVLHSHHSHVTAGHRGGHRGQKMTFA 67
C R F E L+ ++ + ++ VP+ E VLH HS AGH G + T
Sbjct 605 CER---FMISENILYRQIGSVKQLIVPK--EARELVLHLSHSIPWAGHLGVRK----TIE 655
Query 68 ALSKHYYWPGMRAYTTAYVESCTHCRASKSINQKPADLLQQLLI---PFRRWAHVSVDFI 124
+ K++YWPG++A Y +SC C+ S++ P LQ L + PF R + +D +
Sbjct 656 RIKKYFYWPGLKADVVQYCKSCPECQLV-SLHHPPRVPLQPLPLISTPFER---LGMDIV 711
Query 125 TDLPLTTTGHDSILVMVDSLSKMAHFVPAKKSFTAADTVELLADRLIRYHGFPAVLISDR 184
+ ++ G+ +LV+ D ++ P K S A + L R G P +++D+
Sbjct 712 GPVEKSSLGNRFLLVITDYATRYPEVFPLK-SMKAKNVATCLIQFCSRV-GLPREILTDQ 769
Query 185 GPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQTERVNRTLEQMLRTYI 233
G F S L Q+ + IK ++ YHP++DG TER N+TL+QMLR ++
Sbjct 770 GTNFMSTLLKQVYQLLGIKSLRTTPYHPQTDGLTERFNQTLKQMLRKFV 818
> dre:100332169 LReO_3-like
Length=1775
Score = 102 bits (255), Expect = 8e-22, Method: Compositional matrix adjust.
Identities = 61/219 (27%), Positives = 110/219 (50%), Gaps = 9/219 (4%)
Query 18 EPYLHIRVHGLWRICVPQFPEFLTHVLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPG 77
+P ++ V + ++ +P + VL H H AGH G + T+ + K+++WPG
Sbjct 888 KPQVNGDVLAVHQVVLPS--AYRPQVLKLAHEHPLAGHLGITK----TYKRILKYFFWPG 941
Query 78 MRAYTTAYVESCTHCRASKSINQK-PADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDS 136
++ +Y +C C+ S NQ P LQ + + + + +D + LP + +GH
Sbjct 942 LKNSVVSYCRACHDCQLSGKPNQTVPTAPLQPIPVINEPFERLILDCVGPLPKSKSGHQY 1001
Query 137 ILVMVDSLSKMAHFVPAKKSFTAADTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQL 196
IL ++ + ++ P + + E++ + G P V+ +D+G F S L+ Q+
Sbjct 1002 ILTLMCAATRFPEAFPLRSLRASVIVKEII--KFCSIFGLPKVIQTDQGSNFTSKLFAQV 1059
Query 197 CRRFNIKRCMSSSYHPESDGQTERVNRTLEQMLRTYIQS 235
+ + MSS+YHPES G ER ++TL+ MLRT+ S
Sbjct 1060 LKELGVSHQMSSAYHPESQGALERFHQTLKTMLRTFCVS 1098
> dre:100332677 LReO_3-like
Length=1288
Score = 102 bits (255), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 61/196 (31%), Positives = 99/196 (50%), Gaps = 11/196 (5%)
Query 43 VLHSHHSHVTAGHRGGHRGQKMTFAALSKHYYWPGMRAYTTAYVESCTHCR--ASKSINQ 100
VL H+H AGH G T + ++WPG+ Y ++C C+ + +
Sbjct 374 VLELAHTHPMAGHLGAAN----TVKRIRDRFHWPGLDGEVKRYCQACDICQRTSPQRPPP 429
Query 101 KPADLLQQLLIPFRRWAHVSVDFITDLPLTTTGHDSILVMVDSLSKMAHFVPAKKSFTAA 160
P L + +PF R + +D + LP + GH+ ILV++D ++ +P +K+ ++A
Sbjct 430 SPLIPLPIIDVPFTR---IGMDLVGPLPKSARGHEHILVILDYATRYPEAIPLRKATSSA 486
Query 161 DTVELLADRLIRYHGFPAVLISDRGPRFQSDLWNQLCRRFNIKRCMSSSYHPESDGQTER 220
EL L G P +++D+G F S L LC +K+ +S YHP++DG ER
Sbjct 487 IAKELFL--LCSRVGIPTEILTDQGTPFMSRLMADLCHLLKVKQLKTSVYHPQTDGLVER 544
Query 221 VNRTLEQMLRTYIQSD 236
N+TL+QMLR + D
Sbjct 545 FNKTLKQMLRRVVAED 560
Lambda K H
0.326 0.136 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8145948148
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40